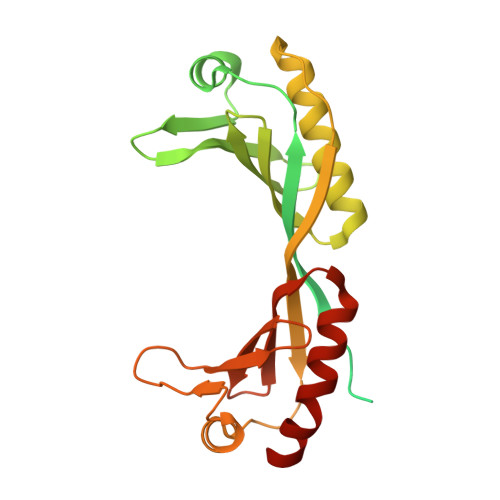
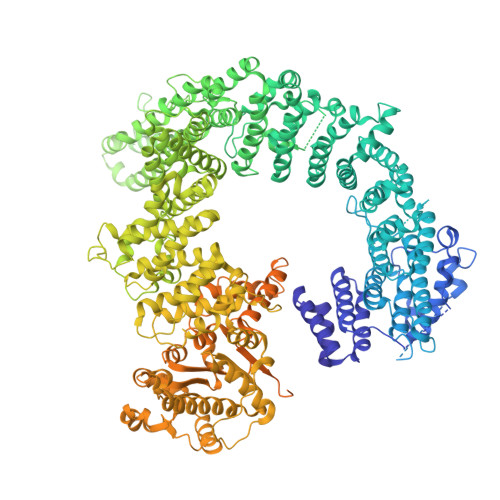
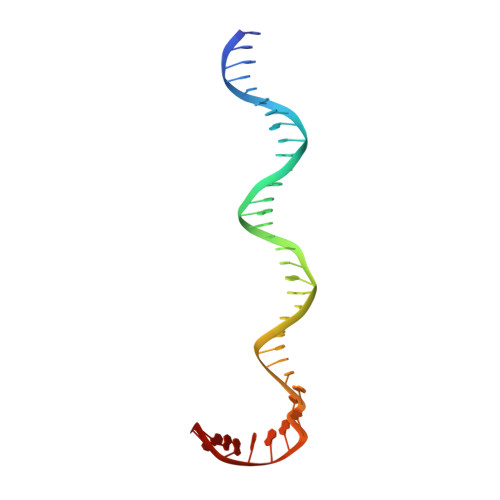
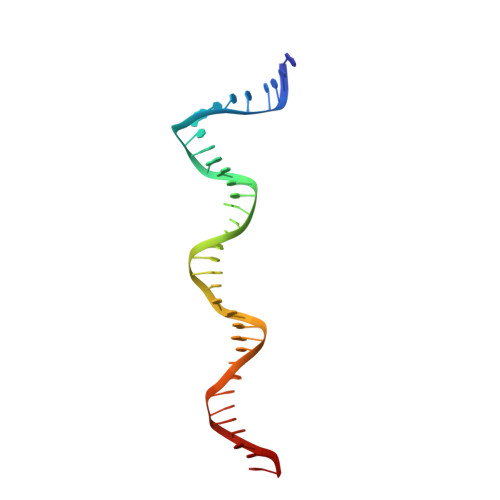
Structural basis for TBP displacement from TATA box DNA by the Swi2/Snf2 ATPase Mot1.
Woike, S., Eustermann, S., Jung, J., Wenzl, S.J., Hagemann, G., Bartho, J., Lammens, K., Butryn, A., Herzog, F., Hopfner, K.P.(2023) Nat Struct Mol Biol 30: 640-649
- PubMed: 37106137
- DOI: https://doi.org/10.1038/s41594-023-00966-0
- Primary Citation of Related Structures:
7Z7N, 7Z8S, 7ZB5, 7ZKE - PubMed Abstract:
The Swi2/Snf2 family transcription regulator Modifier of Transcription 1 (Mot1) uses adenosine triphosphate (ATP) to dissociate and reallocate the TATA box-binding protein (TBP) from and between promoters. To reveal how Mot1 removes TBP from TATA box DNA, we determined cryogenic electron microscopy structures that capture different states of the remodeling reaction. The resulting molecular video reveals how Mot1 dissociates TBP in a process that, intriguingly, does not require DNA groove tracking. Instead, the motor grips DNA in the presence of ATP and swings back after ATP hydrolysis, moving TBP to a thermodynamically less stable position on DNA. Dislodged TBP is trapped by a chaperone element that blocks TBP's DNA binding site. Our results show how Swi2/Snf2 proteins can remodel protein-DNA complexes through DNA bending without processive DNA tracking and reveal mechanistic similarities to RNA gripping DEAD box helicases and RIG-I-like immune sensors.
- Department of Biochemistry, Ludwig-Maximilians-Universität, Munich, Germany.
Organizational Affiliation: