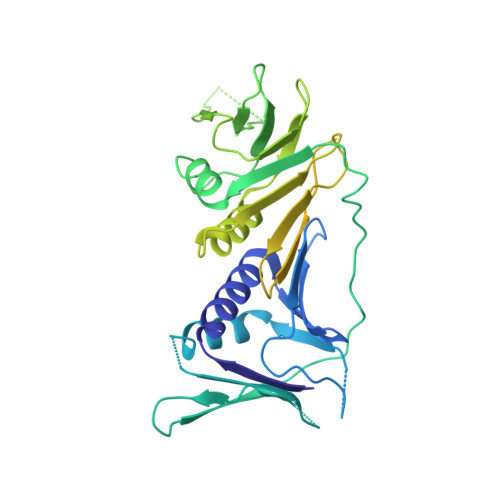
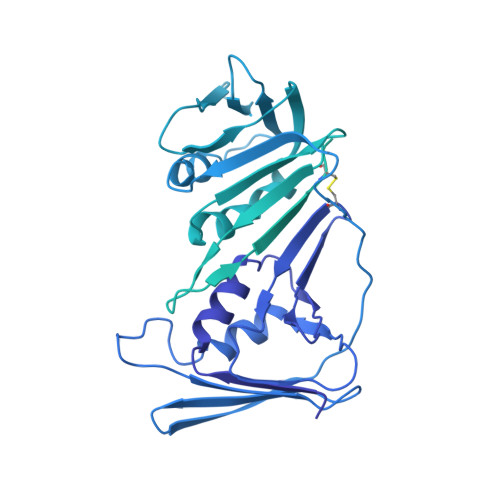
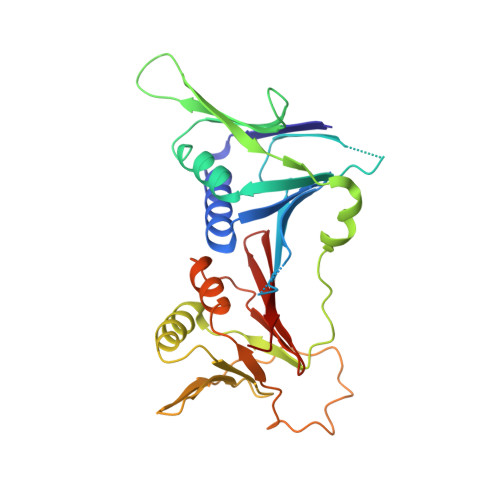
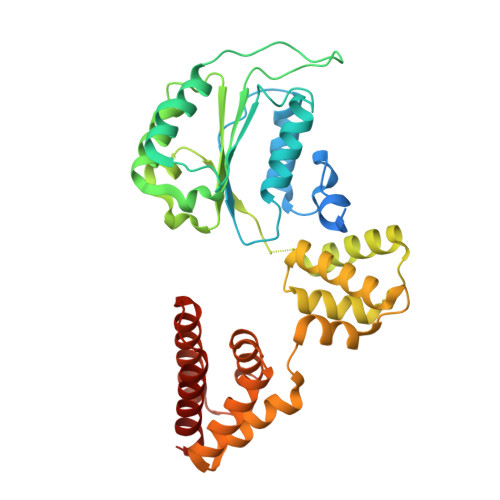
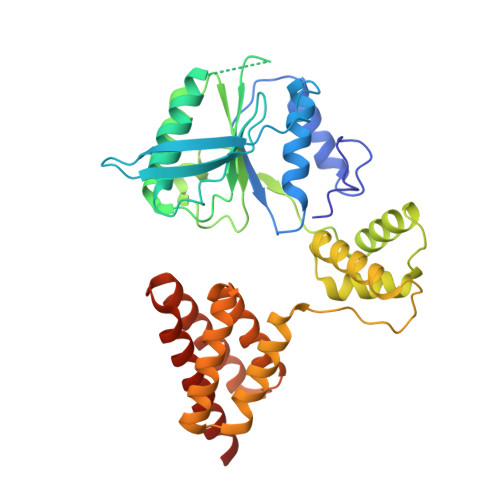
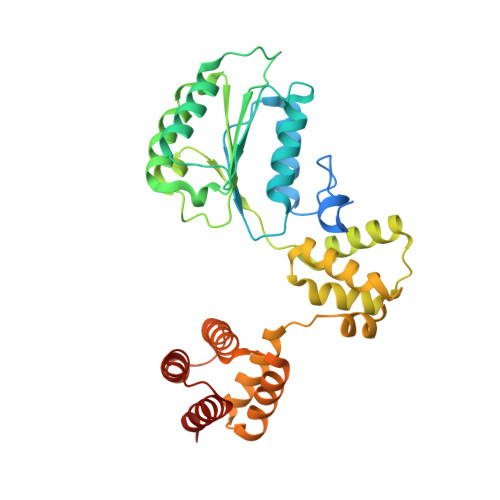
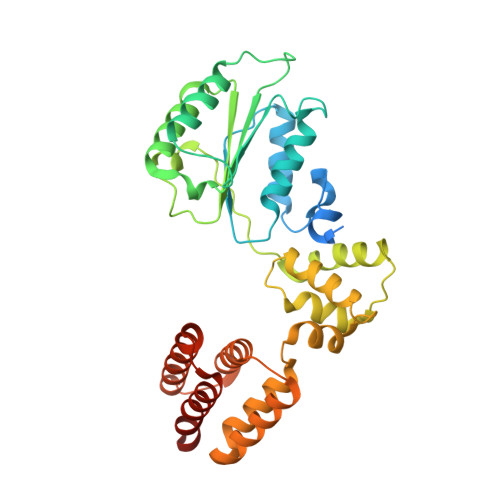
Structure of the human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp bound to a dsDNA-ssDNA junction.
Day, M., Oliver, A.W., Pearl, L.H.(2022) Nucleic Acids Res 50: 8279-8289
- PubMed: 35819203
- DOI: https://doi.org/10.1093/nar/gkac588
- Primary Citation of Related Structures:
7Z6H - PubMed Abstract:
The RAD9-RAD1-HUS1 (9-1-1) clamp forms one half of the DNA damage checkpoint system that signals the presence of substantial regions of single-stranded DNA arising from replication fork collapse or resection of DNA double strand breaks. Loaded at the 5'-recessed end of a dsDNA-ssDNA junction by the RAD17-RFC clamp loader complex, the phosphorylated C-terminal tail of the RAD9 subunit of 9-1-1 engages with the mediator scaffold TOPBP1 which in turn activates the ATR kinase, localised through the interaction of its constitutive partner ATRIP with RPA-coated ssDNA. Using cryogenic electron microscopy (cryoEM) we have determined the structure of a complex of the human RAD17-RFC clamp loader bound to human 9-1-1, engaged with a dsDNA-ssDNA junction. The structure answers the key questions of how RAD17 confers specificity for 9-1-1 over PCNA, and how the clamp loader specifically recognises the recessed 5' DNA end and fixes the orientation of 9-1-1 on the ssDNA.
- Cancer Research UK DNA Repair Enzymes Group, Genome Damage and Stability Centre, School of Life Sciences, University of Sussex, Falmer, Brighton BN1 9RQ, UK.
Organizational Affiliation: