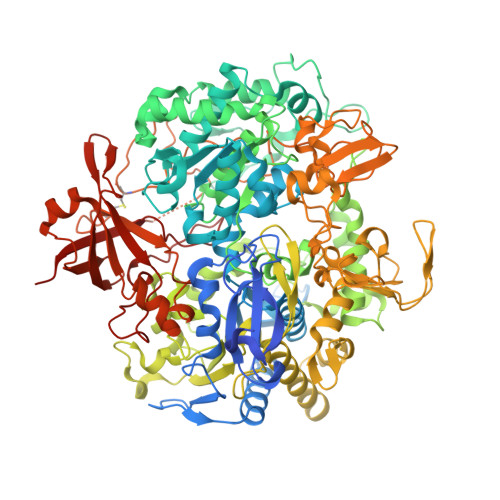
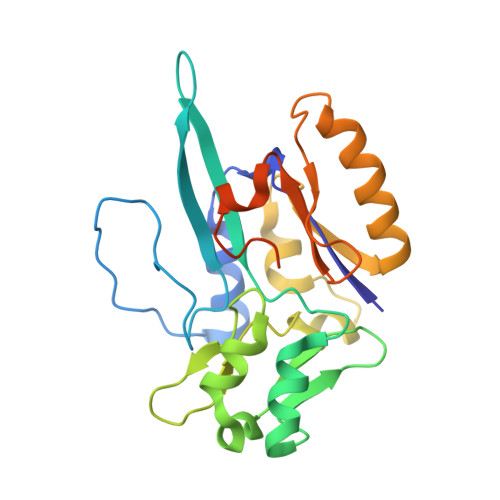
Spectroscopic and Structural Characterization of Reduced Desulfovibrio vulgaris Hildenborough W-FdhAB Reveals Stable Metal Coordination during Catalysis.
Oliveira, A.R., Mota, C., Klymanska, K., Biaso, F., Romao, M.J., Guigliarelli, B., Pereira, I.C.(2022) ACS Chem Biol 17: 1901-1909
- PubMed: 35766974
- DOI: https://doi.org/10.1021/acschembio.2c00336
- Primary Citation of Related Structures:
7Z5O - PubMed Abstract:
Metal-dependent formate dehydrogenases are important enzymes due to their activity of CO 2 reduction to formate. The tungsten-containing FdhAB formate dehydrogenase from Desulfovibrio vulgaris Hildenborough is a good example displaying high activity, simple composition, and a notable structural and catalytic robustness. Here, we report the first spectroscopic redox characterization of FdhAB metal centers by EPR. Titration with dithionite or formate leads to reduction of three [4Fe-4S] 1+ clusters, and full reduction requires Ti(III)-citrate. The redox potentials of the four [4Fe-4S] 1+ centers range between -250 and -530 mV. Two distinct W V signals were detected, W D V and W F V , which differ in only the g 2 -value. This difference can be explained by small variations in the twist angle of the two pyranopterins, as determined through DFT calculations of model compounds. The redox potential of W VI/V was determined to be -370 mV when reduced by dithionite and -340 mV when reduced by formate. The crystal structure of dithionite-reduced FdhAB was determined at high resolution (1.5 Å), revealing the same structural alterations as reported for the formate-reduced structure. These results corroborate a stable six-ligand W coordination in the catalytic intermediate W V state of FdhAB.
- Instituto de Tecnologia Química e Biológica António Xavier, Universidade Nova de Lisboa, Av. da República, 2780-157 Oeiras, Portugal.
Organizational Affiliation: