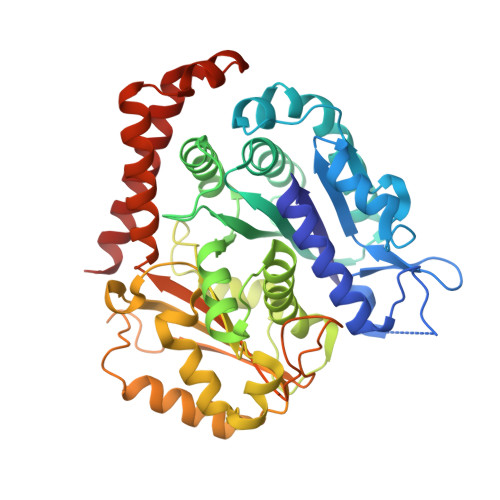
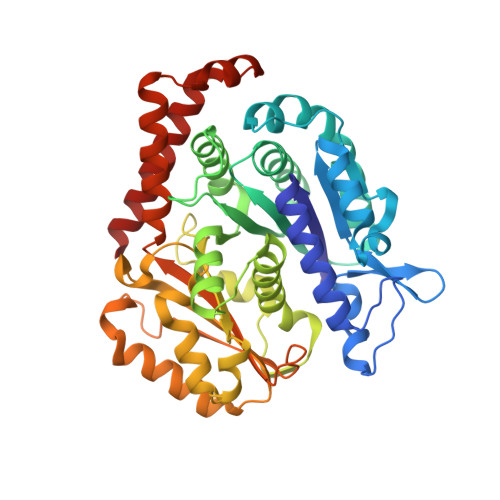
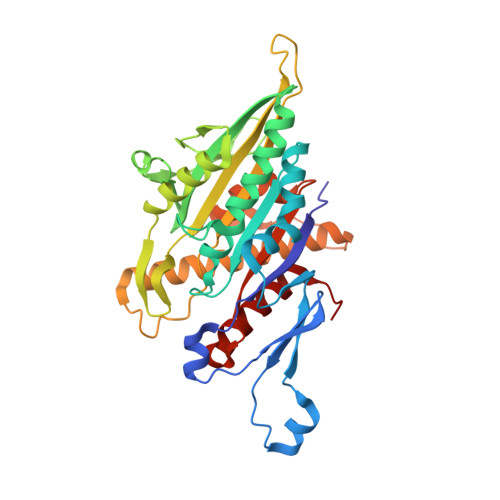
Mechanochemical tuning of a kinesin motor essential for malaria parasite transmission.
Liu, T., Shilliday, F., Cook, A.D., Zeeshan, M., Brady, D., Tewari, R., Sutherland, C.J., Roberts, A.J., Moores, C.A.(2022) Nat Commun 13: 6988-6988
- PubMed: 36384964
- DOI: https://doi.org/10.1038/s41467-022-34710-x
- Primary Citation of Related Structures:
7Z2A, 7Z2B, 7Z2C - PubMed Abstract:
Plasmodium species cause malaria and kill hundreds of thousands annually. The microtubule-based motor kinesin-8B is required for development of the flagellated Plasmodium male gamete, and its absence completely blocks parasite transmission. To understand the molecular basis of kinesin-8B's essential role, we characterised the in vitro properties of kinesin-8B motor domains from P. berghei and P. falciparum. Both motors drive ATP-dependent microtubule gliding, but also catalyse ATP-dependent microtubule depolymerisation. We determined these motors' microtubule-bound structures using cryo-electron microscopy, which showed very similar modes of microtubule interaction in which Plasmodium-distinct sequences at the microtubule-kinesin interface influence motor function. Intriguingly however, P. berghei kinesin-8B exhibits a non-canonical structural response to ATP analogue binding such that neck linker docking is not induced. Nevertheless, the neck linker region is required for motility and depolymerisation activities of these motors. These data suggest that the mechanochemistry of Plasmodium kinesin-8Bs is functionally tuned to support flagella formation.
- Institute of Structural and Molecular Biology, Birkbeck College, London, WC1E 7HX, UK.
Organizational Affiliation: