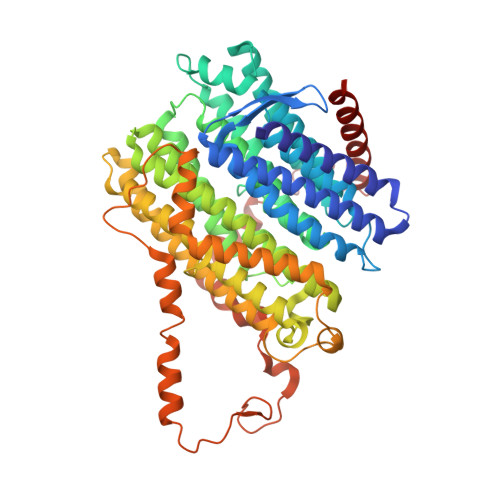
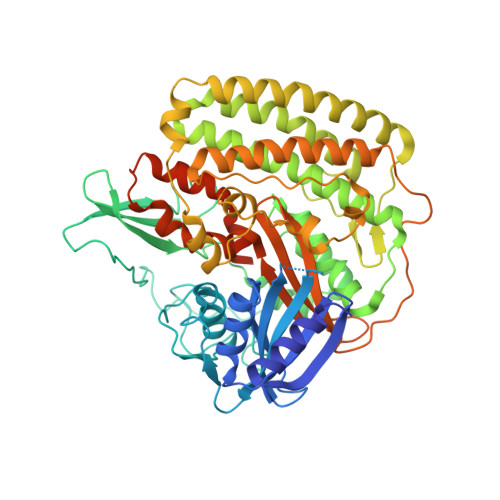
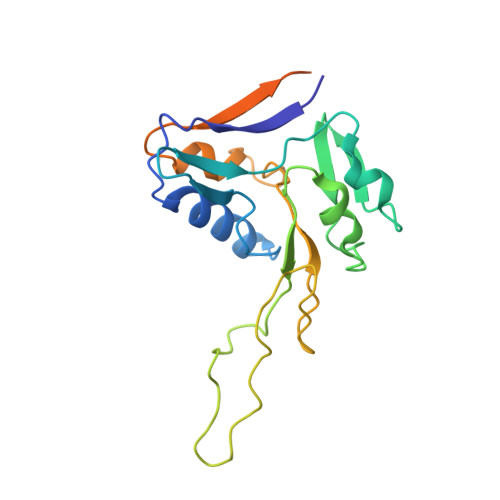
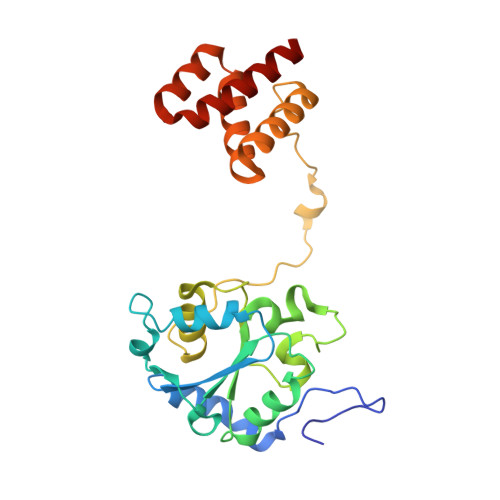
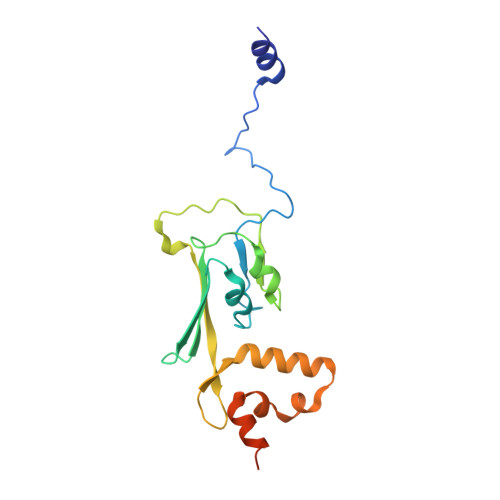
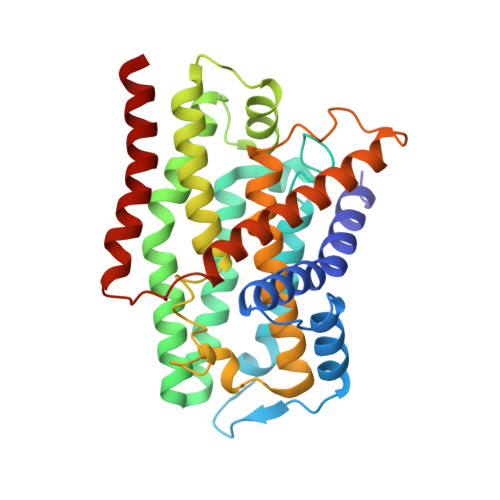
Structure of the membrane-bound formate hydrogenlyase complex from Escherichia coli.
Steinhilper, R., Hoff, G., Heider, J., Murphy, B.J.(2022) Nat Commun 13: 5395-5395
- PubMed: 36104349
- DOI: https://doi.org/10.1038/s41467-022-32831-x
- Primary Citation of Related Structures:
7Z0S, 7Z0T - PubMed Abstract:
The prototypical hydrogen-producing enzyme, the membrane-bound formate hydrogenlyase (FHL) complex from Escherichia coli, links formate oxidation at a molybdopterin-containing formate dehydrogenase to proton reduction at a [NiFe] hydrogenase. It is of intense interest due to its ability to efficiently produce H 2 during fermentation, its reversibility, allowing H 2 -dependent CO 2 reduction, and its evolutionary link to respiratory complex I. FHL has been studied for over a century, but its atomic structure remains unknown. Here we report cryo-EM structures of FHL in its aerobically and anaerobically isolated forms at resolutions reaching 2.6 Å. This includes well-resolved density for conserved loops linking the soluble and membrane arms believed to be essential in coupling enzymatic turnover to ion translocation across the membrane in the complex I superfamily. We evaluate possible structural determinants of the bias toward hydrogen production over its oxidation and describe an unpredicted metal-binding site near the interface of FdhF and HycF subunits that may play a role in redox-dependent regulation of FdhF interaction with the complex.
- Redox and Metalloprotein Research Group, Max Planck Institute of Biophysics, 60438, Frankfurt am Main, Germany.
Organizational Affiliation: