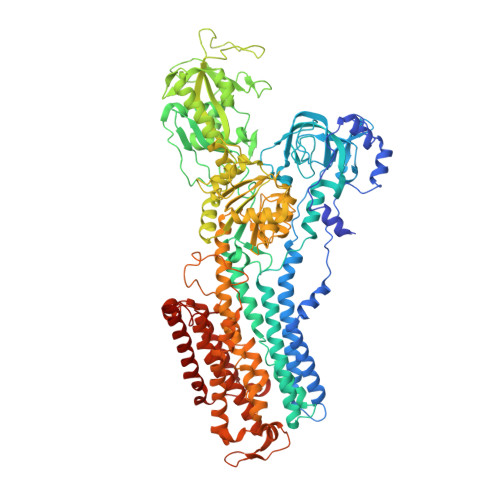
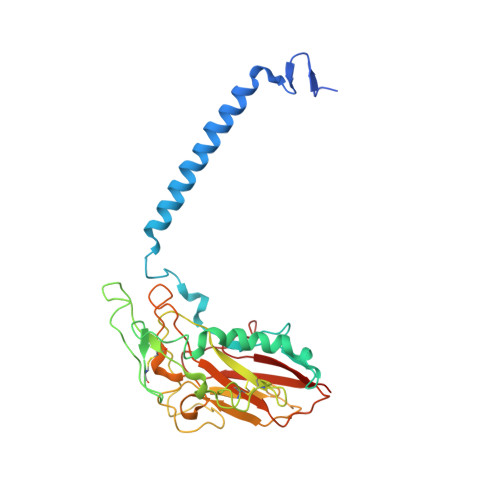
The Na + ,K + -ATPase in complex with beryllium fluoride mimics an ATPase phosphorylated state.
Fruergaard, M.U., Dach, I., Andersen, J.L., Ozol, M., Shahsavar, A., Quistgaard, E.M., Poulsen, H., Fedosova, N.U., Nissen, P.(2022) J Biological Chem 298: 102317-102317
- PubMed: 35926706
- DOI: https://doi.org/10.1016/j.jbc.2022.102317
- Primary Citation of Related Structures:
7QTV, 7YZR, 7Z04 - PubMed Abstract:
The Na + ,K + -ATPase generates electrochemical gradients of Na + and K + across the plasma membrane via a functional cycle that includes various phosphoenzyme intermediates. However, the structure and function of these intermediates and how metal fluorides mimick them require further investigation. Here, we describe a 4.0 Å resolution crystal structure and functional properties of the pig kidney Na + ,K + -ATPase stabilized by the inhibitor beryllium fluoride (denoted E2-BeF x ). E2-BeF x is expected to mimic properties of the E2P phosphoenzyme, yet with unknown characteristics of ion and ligand binding. The structure resembles the E2P form obtained by phosphorylation from inorganic phosphate (P i ) and stabilized by cardiotonic steroids, including a low-affinity Mg 2+ site near ion binding site II. Our anomalous Fourier analysis of the crystals soaked in Rb + (a K + congener) followed by a low-resolution rigid-body refinement (6.9-7.5 Å) revealed preocclusion transitions leading to activation of the dephosphorylation reaction. We show that the Mg 2+ location indicates a site of initial K + recognition and acceptance upon binding to the outward-open E2P state after Na + release. Furthermore, using binding and activity studies, we find that the BeF x -inhibited enzyme is also able to bind ADP/ATP and Na + . These results relate the E2-BeF x complex to a transient K + - and ADP-sensitive E∗P intermediate of the functional cycle of the Na + ,K + -ATPase, prior to E2P.
- Department of Molecular Biology and Genetics, DANDRITE - Nordic EMBL Partnership for Molecular Medicine, Aarhus University, Aarhus C, Denmark.
Organizational Affiliation: