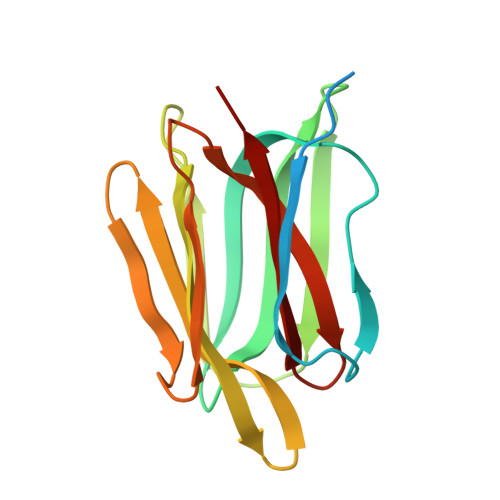
The Crystal Structure of the Defense Conferring Rice Protein Os JAC1 Reveals a Carbohydrate Binding Site on the Dirigent-like Domain.
Huwa, N., Weiergraber, O.H., Fejzagic, A.V., Kirsch, C., Schaffrath, U., Classen, T.(2022) Biomolecules 12
- PubMed: 36009020
- DOI: https://doi.org/10.3390/biom12081126
- Primary Citation of Related Structures:
7R5Z, 7YWE, 7YWF, 7YWG, 7YWW - PubMed Abstract:
Pesticides are routinely used to prevent severe losses in agriculture. This practice is under debate because of its potential negative environmental impact and selection of resistances in pathogens. Therefore, the development of disease resistant plants is mandatory. It was shown that the rice ( Oryza sativa ) protein Os JAC1 enhances resistance against different bacterial and fungal plant pathogens in rice, barley, and wheat. Recently we reported possible carbohydrate interaction partners for both domains of Os JAC1 (a jacalin-related lectin (JRL) and a dirigent (DIR) domain), however, a mechanistic understanding of its function is still lacking. Here, we report crystal structures for both individual domains and the complex of galactobiose with the DIR domain, which revealed a new carbohydrate binding motif for DIR proteins. Docking studies of the two domains led to a model of the full-length protein. Our findings offer insights into structure and binding properties of Os JAC1 and its possible function in pathogen resistance.
- Institute for Bioorganic Chemistry, Heinrich Heine University Düsseldorf, 52425 Jülich, Germany.
Organizational Affiliation: