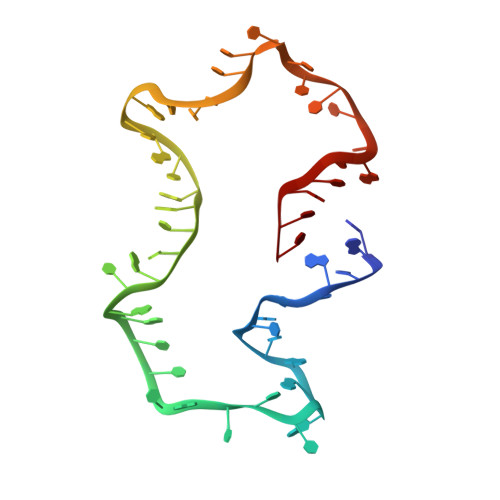
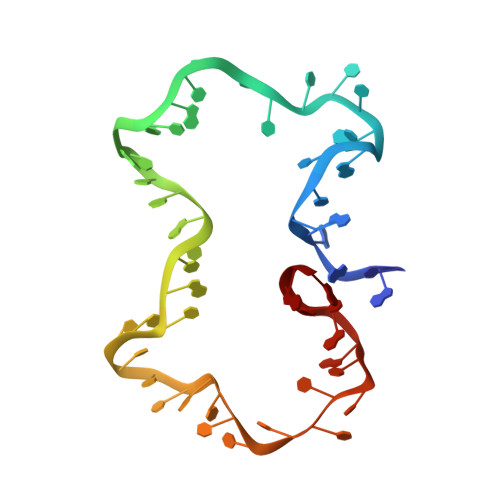
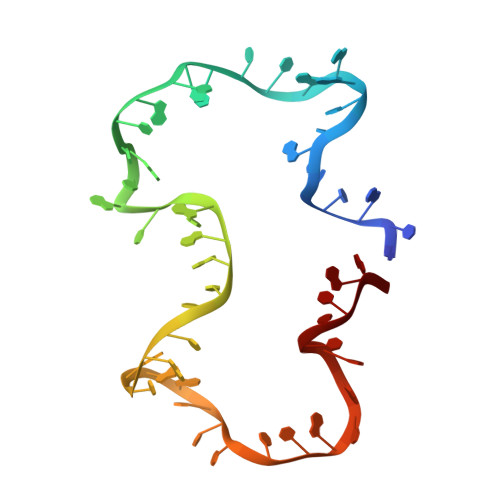
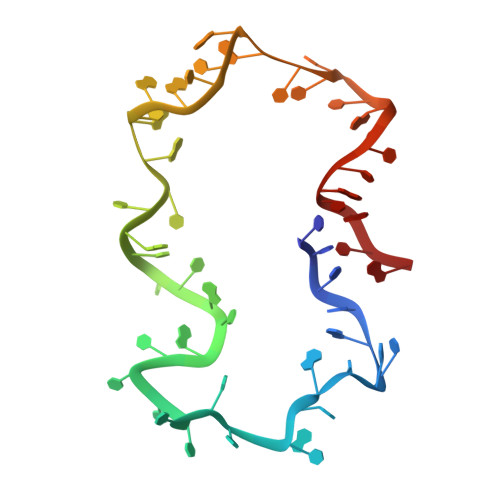
Structure and dynamics of an archetypal DNA nanoarchitecture revealed via cryo-EM and molecular dynamics simulations.
Ahmad, K., Javed, A., Lanphere, C., Coveney, P.V., Orlova, E.V., Howorka, S.(2023) Nat Commun 14: 3630-3630
- PubMed: 37336895
- DOI: https://doi.org/10.1038/s41467-023-38681-5
- Primary Citation of Related Structures:
7YWH, 7YWI, 7YWL, 7YWN, 7YWO - PubMed Abstract:
DNA can be folded into rationally designed, unique, and functional materials. To fully realise the potential of these DNA materials, a fundamental understanding of their structure and dynamics is necessary, both in simple solvents as well as more complex and diverse anisotropic environments. Here we analyse an archetypal six-duplex DNA nanoarchitecture with single-particle cryo-electron microscopy and molecular dynamics simulations in solvents of tunable ionic strength and within the anisotropic environment of biological membranes. Outside lipid bilayers, the six-duplex bundle lacks the designed symmetrical barrel-type architecture. Rather, duplexes are arranged in non-hexagonal fashion and are disorted to form a wider, less elongated structure. Insertion into lipid membranes, however, restores the anticipated barrel shape due to lateral duplex compression by the bilayer. The salt concentration has a drastic impact on the stability of the inserted barrel-shaped DNA nanopore given the tunable electrostatic repulsion between the negatively charged duplexes. By synergistically combining experiments and simulations, we increase fundamental understanding into the environment-dependent structural dynamics of a widely used nanoarchitecture. This insight will pave the way for future engineering and biosensing applications.
- Centre for Computational Science, University College London, London, WC1H 0AJ, UK.
Organizational Affiliation: