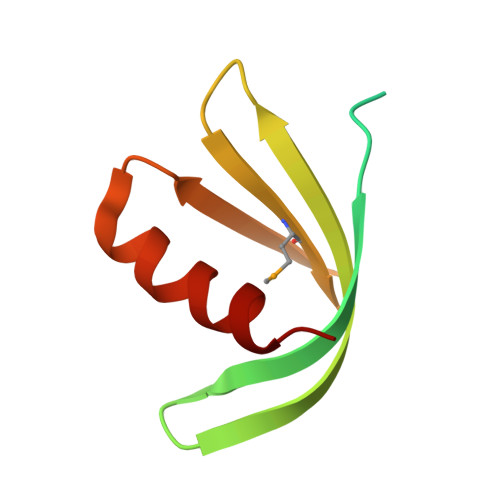
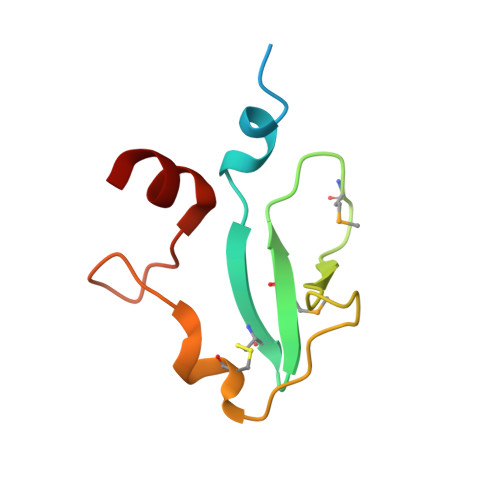
Crystal structure of vaccinia virus G3/L5 sub-complex reveals a novel fold with extended inter-molecule interactions conserved among orthopoxviruses.
Lin, S., Yue, D., Yang, F., Chen, Z., He, B., Cao, Y., Dong, H., Li, J., Zhao, Q., Lu, G.(2023) Emerg Microbes Infect 12: e2160661-e2160661
- PubMed: 36533407
- DOI: https://doi.org/10.1080/22221751.2022.2160661
- Primary Citation of Related Structures:
7YTT, 7YTU - West China Hospital Emergency Department (WCHED), State Key Laboratory of Biotherapy, West China Hospital, Sichuan University, Chengdu, People's Republic of China.
Organizational Affiliation: