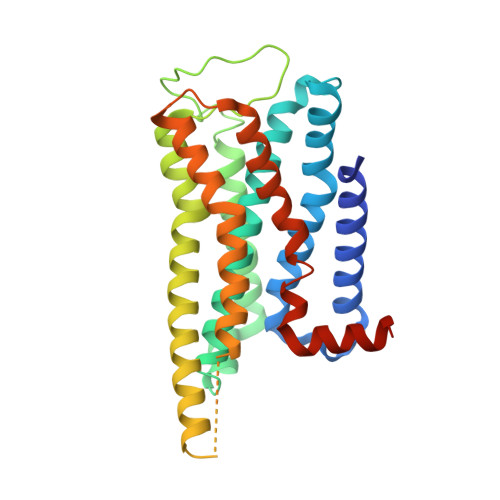
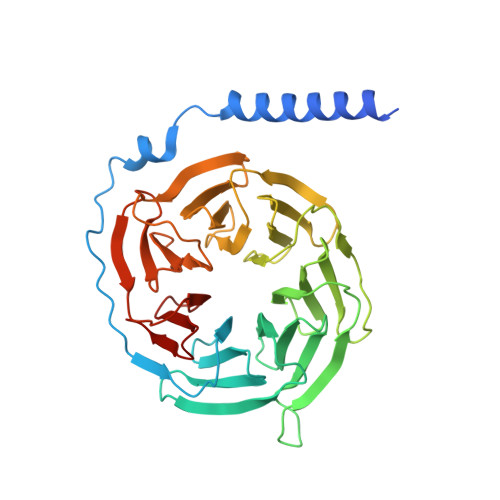
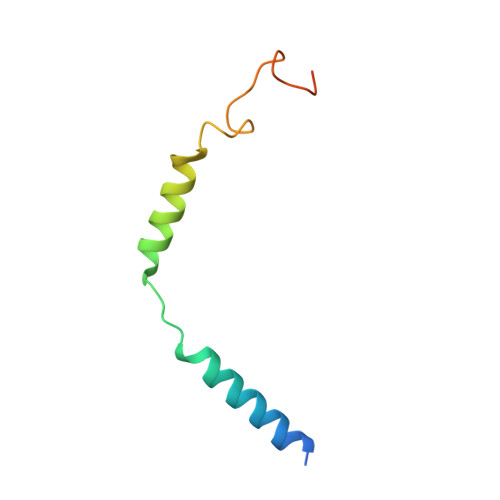
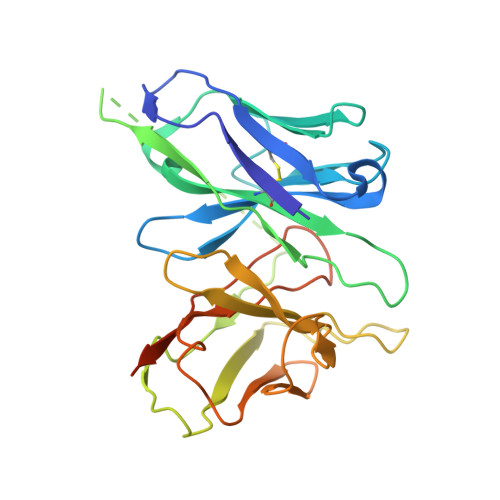
Structural insights into constitutive activity of 5-HT 6 receptor.
He, L., Zhao, Q.Y., Qi, J., Wang, Y., Han, W., Chen, Z., Cong, Y., Wang, S.(2023) Proc Natl Acad Sci U S A 120: e2209917120-e2209917120
- PubMed: 36989299
- DOI: https://doi.org/10.1073/pnas.2209917120
- Primary Citation of Related Structures:
7YS6 - PubMed Abstract:
While most therapeutic research on G-protein-coupled receptors (GPCRs) focuses on receptor activation by (endogenous) agonists, significant therapeutic potential exists through agonist-independent intrinsic constitutive activity that can occur in various physiological and pathophysiological settings. For example, inhibiting the constitutive activity of 5-HT 6 R-a receptor that is found almost exclusively in the brain and mediates excitatory neurotransmission-has demonstrated a therapeutic effect on cognitive/memory impairment associated with several neuropsychiatric disorders. However, the structural basis of such constitutive activity remains unclear. Here, we present a cryo-EM structure of serotonin-bound human 5-HT 6 R-Gs heterotrimer at 3.0-Å resolution. Detailed analyses of the structure complemented by comprehensive interrogation of signaling illuminate key structural determinants essential for constitutive 5-HT 6 R activity. Additional structure-guided mutagenesis leads to a nanobody mimic Gαs for 5-HT 6 R that can reduce its constitutive activity. Given the importance of 5-HT 6 R for a large number of neuropsychiatric disorders, insights derived from these studies will accelerate the design of more effective medications, and shed light on the molecular basis of constitutive activity.
- State Key Laboratory of Molecular Biology, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, University of Chinese Academy of Sciences, Shanghai 200031, China.
Organizational Affiliation: