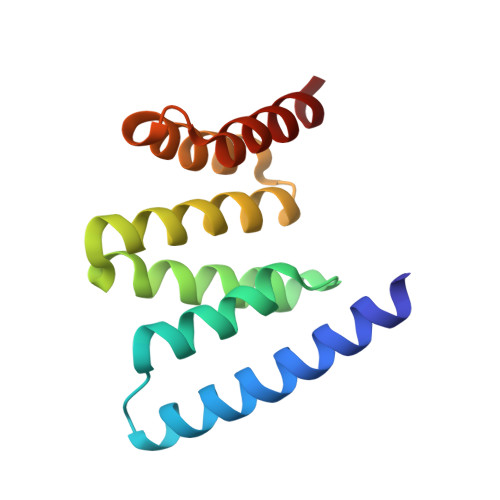
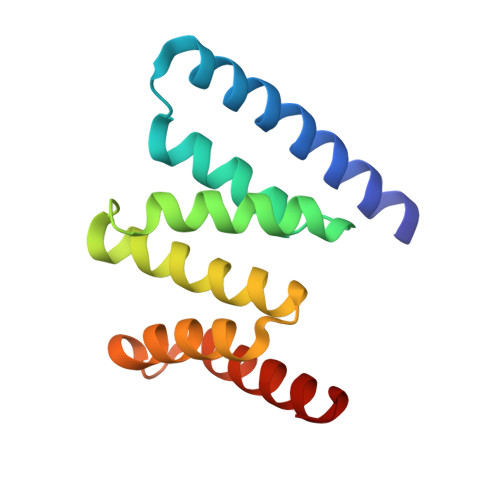
Crystal structure of Fis1 and Bap31 provides information on protein-protein interactions at mitochondria-associated ER membranes
Nguyen, M.D., Kim, Y., Bae, S.H., Kim, S., Yeo, H.K., Ha, N.C., Cho, G., Moon, S., Cho, K.H., Jang, H., Bong, S.M., Lee, B.I.(2025) Commun Biol 8: 1161