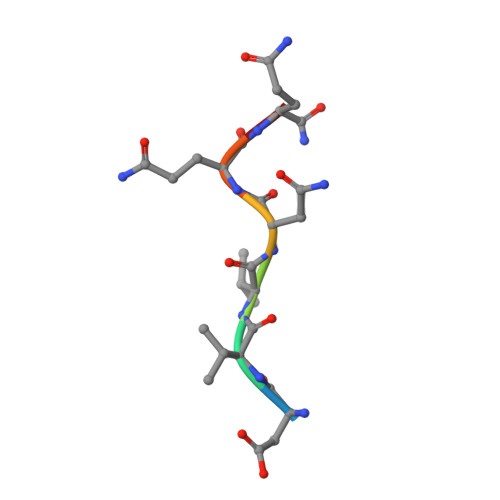
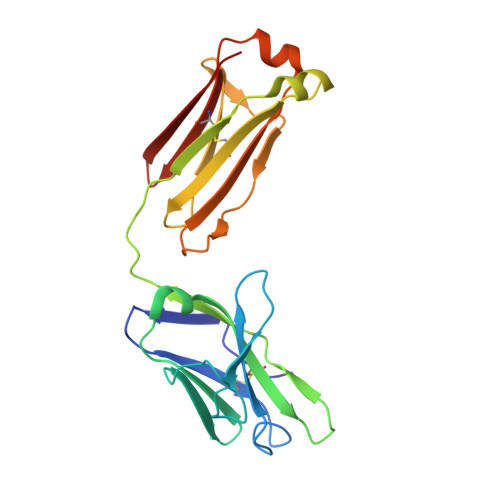
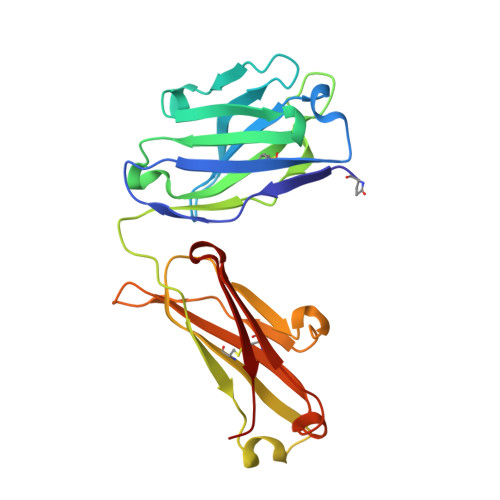
A broadly neutralizing antibody recognizes a unique epitope with a signature motif common across coronaviruses.
Yan, L., Wang, F., Hill, M., Brun, J., Liang, Z., Shi, X., Zhang, L., He, X., Li, Y., Huang, Q., Dong, X., Liu, H., Zhang, Y., Liu, L., Dwek, R.A., Zitzmann, N., Liang, A., Yang, G.(2025) Nat Commun 16: 7580-7580
- PubMed: 40813592
- DOI: https://doi.org/10.1038/s41467-025-63101-1
- Primary Citation of Related Structures:
7Y8J, 7YI6 - PubMed Abstract:
Cross-reactive antibodies targeting multiple epitopes have been identified in Sarbecoviruses, but the precise molecular mechanism(s) behind the crossreactivity remain poorly understood. Here, we isolate 3D1, a broadly neutralizing antibody (bnAb) derived from a human combinatorial antibody library targeting the conserved HR1 domain. 3D1 uniquely recognizes a β-turn fold comprising a 6-mer peptide (pep DVVNQN/Q ) that forms during a pre-hairpin transition state, occurring exclusively before membrane fusion during viral infection. 3D1 effectively neutralizes a wide range of live SARS-CoV-2 wild-type strains except for Omicron, which evades neutralization due to a detrimental point mutation (Q954H). Notably, this cryptic epitope reveals a signature motif that extends throughout the core region of coronaviruses and is also present in various RNA viruses, including HIV and Marburgvirus. 3D1 functions as a natural or background antibody capable of binding to a diverse array of non-self antigens. 3D1's cross-reactivity underscores the effectiveness of the library approach, which encompasses the entire antibody repertoire.
- Shanghai Institute for Advanced Immunochemical Studies, ShanghaiTech University, Shanghai, China.
Organizational Affiliation: