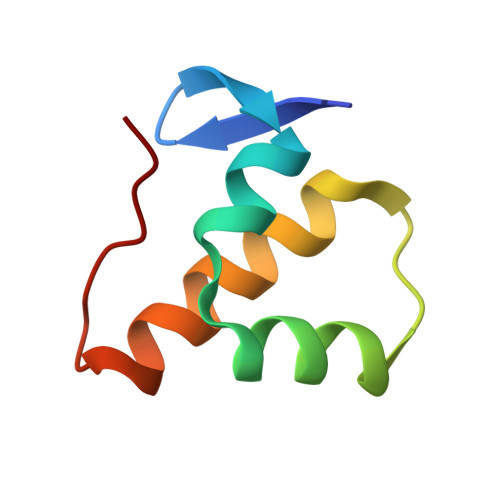
High-resolution crystal structure of the anti-CRISPR protein AcrIC5.
Kang, Y.J., Park, H.H.(2022) Biochem Biophys Res Commun 625: 102-108
- PubMed: 35952606
- DOI: https://doi.org/10.1016/j.bbrc.2022.08.005
- Primary Citation of Related Structures:
7YHR - PubMed Abstract:
As a result of the long-term battle of bacteria and archaea against invaders such as viruses and genetic mobile elements, they have developed CRISPR-Cas systems for self-defense, which allows them to remove the viral genetic material introduced into host cells via infection. To fight against this bacterial immune system, however, viruses have also evolved to produce multiple anti-CRISPR proteins that can inhibit the bacterial CRISPR-Cas system. In this study, we introduced a tentative inhibitory activity against a type I-C CRISPR-Cas system by determining the crystal structure of AcrIC5 from Pseudomonas delhiensis. Structural analysis revealed that AcrIC5 was composed of noble folds comprising two antiparallel sheets and three helices. Although AcrIC5 did not directly interact with either the type I-C cascade from Neisseria lactamia or the type I-F cascade from Pseudomonas aeruginosa in our analysis, a highly acidic surface feature indicated that AcrIC5 may be DNA mimic Acrs that directly binds to the target DNA binding site in type I-C cascade and inhibits the recruitment of the target DNA to this cascade.
- College of Pharmacy, Chung-Ang University, Seoul, 06974, Republic of Korea; Department of Global Innovative Drugs, Graduate School of Chung-Ang University, Seoul, 06974, Republic of Korea.
Organizational Affiliation: