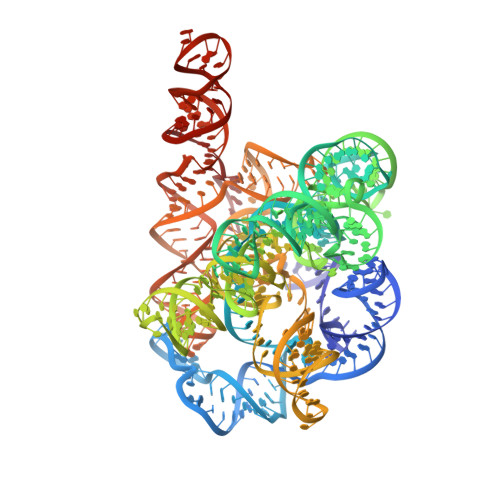
Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Li, S., Palo, M.Z., Zhang, X., Pintilie, G., Zhang, K.(2023) Nat Commun 14: 1294-1294
- PubMed: 36928031
- DOI: https://doi.org/10.1038/s41467-023-36724-5
- Primary Citation of Related Structures:
7YG8, 7YG9, 7YGA, 7YGB, 7YGC, 7YGD - PubMed Abstract:
Group I introns are catalytic RNAs that coordinate two consecutive transesterification reactions for self-splicing. To understand how the group I intron promotes catalysis and coordinates self-splicing reactions, we determine the structures of L-16 Tetrahymena ribozyme in complex with a 5'-splice site analog product and a 3'-splice site analog substrate using cryo-EM. We solve six conformations from a single specimen, corresponding to different splicing intermediates after the first ester-transfer reaction. The structures reveal dynamics during self-splicing, including large conformational changes of the internal guide sequence and the J5/4 junction as well as subtle rearrangements of active-site metals and the hydrogen bond formed between the 2'-OH group of A261 and the N2 group of guanosine substrate. These results help complete a detailed structural and mechanistic view of this paradigmatic group I intron undergoing the second step of self-splicing.
- Department of Urology, The First Affiliated Hospital of USTC, MOE Key Laboratory for Cellular Dynamics, Hefei National Research Center for Interdisciplinary Sciences at the Microscale, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, 230001, China. lishanshan@ustc.edu.cn.
Organizational Affiliation: