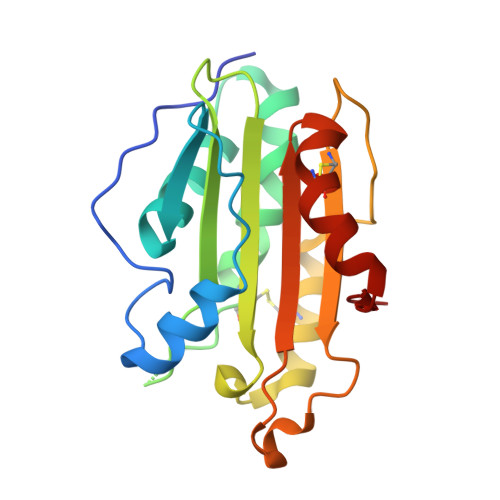
Crystal structure of a mycobacterial secretory protein Rv0398c and in silico prediction of its export pathway.
Saha, R., Mukherjee, S., Singh, B., De, S., Weiss, M.S., Das, A.K.(2023) Biochem Biophys Res Commun 672: 45-53
- PubMed: 37336124
- DOI: https://doi.org/10.1016/j.bbrc.2023.06.029
- Primary Citation of Related Structures:
7YD4 - PubMed Abstract:
Secretory proteins are used by pathogenic bacteria to manipulate the host systems and compete with other microorganisms, thereby enabling their survival in their host. Similar to other bacteria, secretory proteins of Mycobacterium tuberculosis also play a pivotal role in evading immune response within hosts, thereby leading to acute and latent tuberculosis infection. Prokaryotes have several classes of bacterial secretory systems out of which the Sec and Tat pathways are the most conserved in Mtb to transport proteins across the cytoplasmic membrane. Here, we report the crystal structure of a secretory protein, Rv0398c determined to 1.9 Å resolution. The protein comprises a core of antiparallel β sheets surrounded by α helices adopting a unique β sandwich fold. Structural comparison with other secretory proteins in Mtb and other pathogenic bacteria reveals that Rv0398c may be secreted via the Sec pathway. Our structural and in silico analyses thus provide mechanistic insights into the pathway adopted by Mtb to transport out secretory protein, Rv0398c which will facilitate the invasion to the host immune system.
- Department of Biotechnology, Indian Institute of Technology Kharagpur, Kharagpur, 721302, India.
Organizational Affiliation: