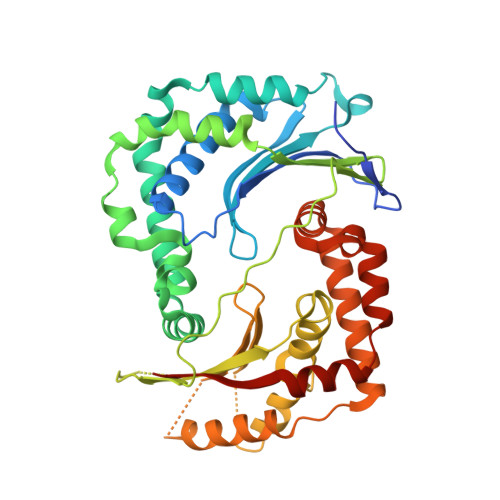
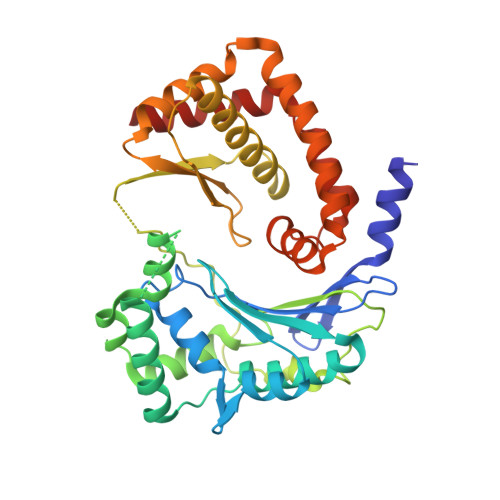
Crystal structure of the AlbEF complex involved in subtilosin A biosynthesis.
Ishida, K., Nakamura, A., Kojima, S.(2022) Structure 30: 1637-1646.e3
- PubMed: 36302388
- DOI: https://doi.org/10.1016/j.str.2022.10.002
- Primary Citation of Related Structures:
7Y8U, 7Y8V, 7Y8X - PubMed Abstract:
Subtilosin A is a sactipeptide bacteriocin produced by Bacillus subtilis strain 168, containing intramolecular thioether bonds and a head-to-tail macrocyclic peptide bond. Macrocyclization is presumably catalyzed by AlbE and AlbF proteins encoded by the subtilosin A biosynthesis gene cluster. However, the underlying mechanism of macrocyclization remains uncertain as the tertiary structures of the proteins are undetermined. Here, we report the crystal structures of AlbE and AlbF homologs in Quasibacillus thermotolerans, wherein the subtilosin biosynthesis gene cluster is highly conserved. Structural analysis and pull-down assays revealed that AlbE and AlbF form heterodimeric complexes. Although the AlbEF complex shows structural similarity to M16B family metalloproteases, the substrate-binding chamber is shallower and more open than the other M16B family proteins. The chamber surface showed electrostatic complementarity to the precursor of subtilosin. Our findings provide insights into the role of AlbEF in metalloprotease catalysis and macrocyclic peptide bond formation.
- Department of Life Science, Graduate School of Science, Gakushuin University, 1-5-1 Mejiro, Toshima-ku, Tokyo 171-8588, Japan.
Organizational Affiliation: