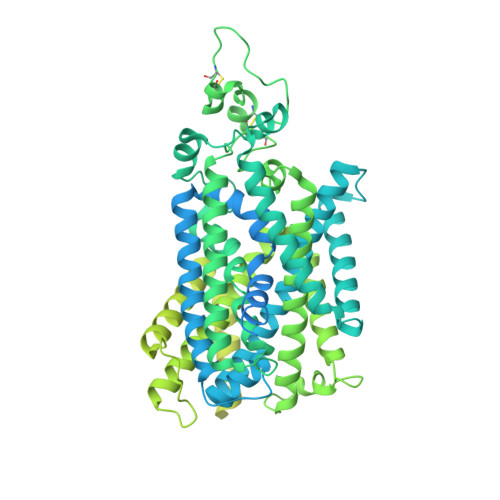
Cryo-EM structure of the human sodium-chloride cotransporter NCC.
Nan, J., Yuan, Y., Yang, X., Shan, Z., Liu, H., Wei, F., Zhang, W., Zhang, Y.(2022) Sci Adv 8: eadd7176-eadd7176
- PubMed: 36351028
- DOI: https://doi.org/10.1126/sciadv.add7176
- Primary Citation of Related Structures:
7Y6I, 7YG0, 7YG1 - PubMed Abstract:
The sodium-chloride cotransporter NCC mediates the coupled import of sodium and chloride across the plasma membrane, playing vital roles in kidney extracellular fluid volume and blood pressure control. Here, we present the full-length structure of human NCC, with 2.9 Å for the transmembrane domain and 3.8 Å for the carboxyl-terminal domain. NCC adopts an inward-open conformation and a domain-swap dimeric assembly. Conserved ion binding sites among the cation-chloride cotransporters and the Na2 site are observed in our structure. A unique His residue in the substrate pocket in NCC potentially interacts with Na1 and Cl1 and might also mediate the coordination of Na2 through a Ser residue. Putative observed water molecules are indicated to participate in the coordination of ions and TM coupling. Together with transport activity assays, our structure provides the first glimpse of NCC and defines ion binding sites, promoting drug development for hypertension targeting on NCC.
- Shanghai Fifth People's Hospital, Fudan University, and Shanghai Key Laboratory of Medical Epigenetics, International Co-laboratory of Medical Epigenetics and Metabolism (Ministry of Science and Technology), Institutes of Biomedical Sciences, Fudan University, Shanghai 200032, China.
Organizational Affiliation: