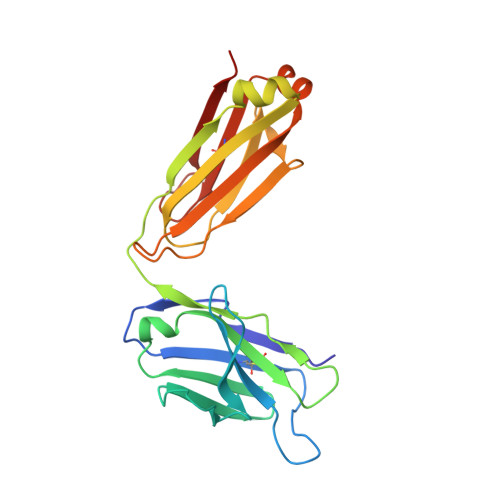
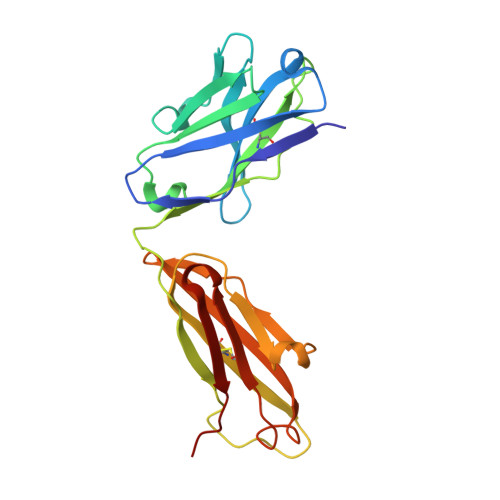
Structural basis of the 24B3 antibody against the toxic conformer of amyloid beta with a turn at positions 22 and 23.
Irie, Y., Matsushima, Y., Kita, A., Miki, K., Segawa, T., Maeda, M., Yanagita, R.C., Irie, K.(2022) Biochem Biophys Res Commun 621: 162-167
- PubMed: 35839743
- DOI: https://doi.org/10.1016/j.bbrc.2022.07.010
- Primary Citation of Related Structures:
7Y3J - PubMed Abstract:
Amyloid β-protein (Aβ) oligomers are involved in the early stages of Alzheimer's disease (AD) and antibodies against these toxic oligomers could be useful for accurate diagnosis of AD. We identified the toxic conformer of Aβ42 with a turn at positions 22/23, which has a propensity to form toxic oligomers. The antibody 24B3, developed by immunization of a toxic conformer surrogate E22P-Aβ9-35 in mice, was found to be useful for AD diagnosis using human cerebrospinal fluid (CSF). However, it is not known how 24B3 recognizes the toxic conformation of wild-type Aβ in CSF. Here, we report the crystal structure of 24B3 Fab complexed with E22P-Aβ11-34, whose residues 16-26 were observed in electron densities, suggesting that the residues comprising the toxic turn at positions 22/23 were recognized by 24B3. Since 24B3 bound only to Aβ42 aggregates, several conformationally restricted analogs of Aβ42 with an intramolecular disulfide bond to mimic the conformation of toxic Aβ42 aggregates were screened by enzyme immunoassay. As a result, only F19C,A30homoC-SS-Aβ42 (1) bound significantly to 24B3. These data provide a structural basis for its low affinity to the Aβ42 monomer and selectivity for its aggregate form.
- Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Sakyo-Ku, Kyoto, 606-8502, Japan.
Organizational Affiliation: