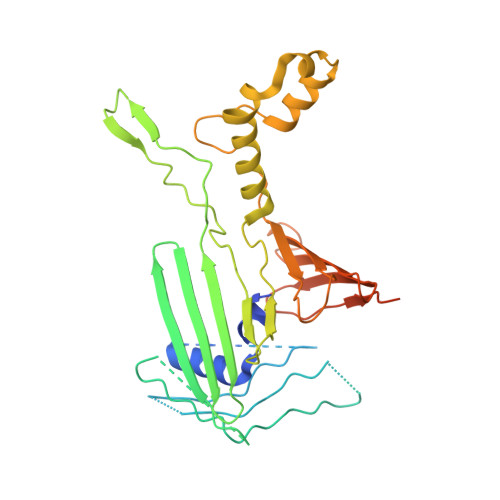
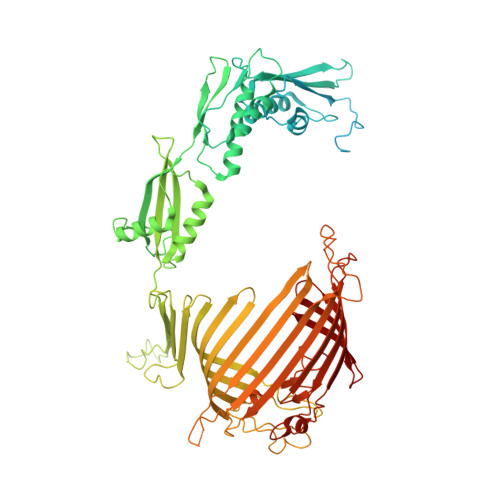
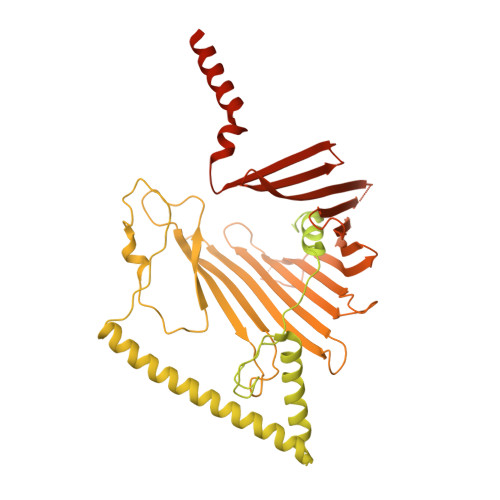
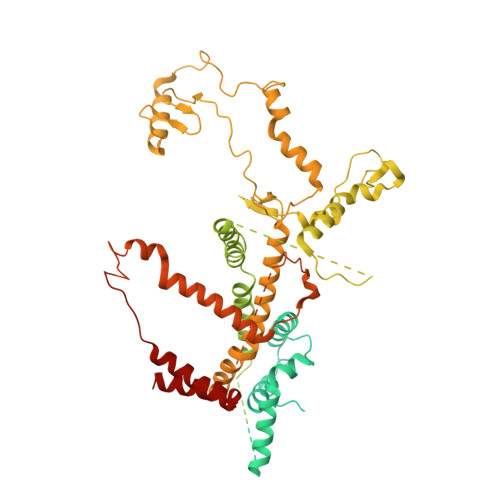
Architecture of chloroplast TOC-TIC translocon supercomplex.
Liu, H., Li, A., Rochaix, J.D., Liu, Z.(2023) Nature 615: 349-357
- PubMed: 36702157
- DOI: https://doi.org/10.1038/s41586-023-05744-y
- Primary Citation of Related Structures:
7XZI, 7XZJ - PubMed Abstract:
Chloroplasts rely on the translocon complexes in the outer and inner envelope membranes (the TOC and TIC complexes, respectively) to import thousands of different nuclear-encoded proteins from the cytosol 1-4 . Although previous studies indicated that the TOC and TIC complexes may assemble into larger supercomplexes 5-7 , the overall architectures of the TOC-TIC supercomplexes and the mechanism of preprotein translocation are unclear. Here we report the cryo-electron microscopy structure of the TOC-TIC supercomplex from Chlamydomonas reinhardtii. The major subunits of the TOC complex (Toc75, Toc90 and Toc34) and TIC complex (Tic214, Tic20, Tic100 and Tic56), three chloroplast translocon-associated proteins (Ctap3, Ctap4 and Ctap5) and three newly identified small inner-membrane proteins (Simp1-3) have been located in the supercomplex. As the largest protein, Tic214 traverses the inner membrane, the intermembrane space and the outer membrane, connecting the TOC complex with the TIC proteins. An inositol hexaphosphate molecule is located at the Tic214-Toc90 interface and stabilizes their assembly. Four lipid molecules are located within or above an inner-membrane funnel formed by Tic214, Tic20, Simp1 and Ctap5. Multiple potential pathways found in the TOC-TIC supercomplex may support translocation of different substrate preproteins into chloroplasts.
- National Laboratory of Biomacromolecules, CAS Centre for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China.
Organizational Affiliation: