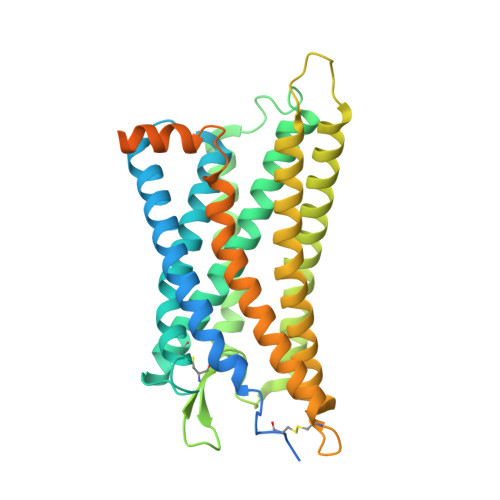
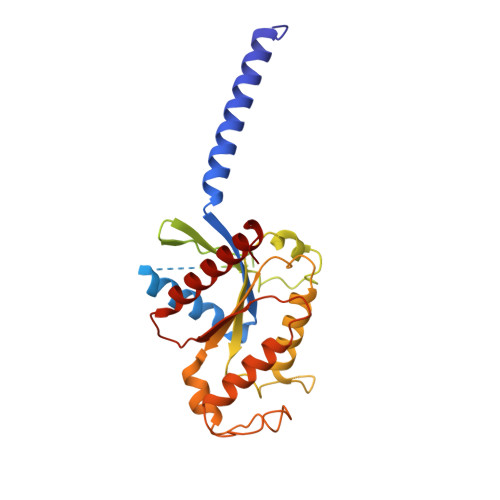
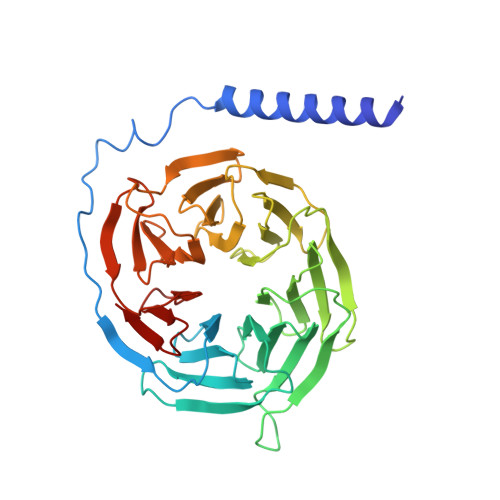
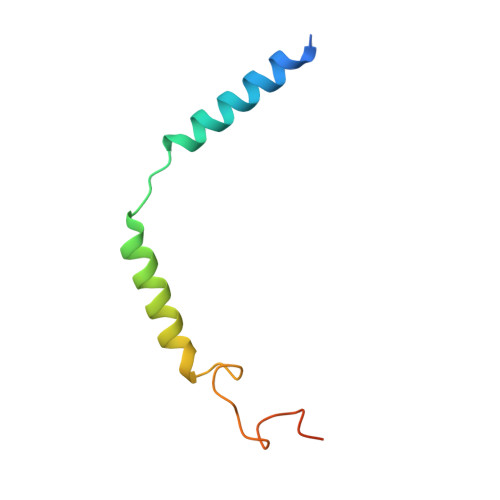
Structural insights into signal transduction of the purinergic receptors P2Y1R and P2Y12R.
Li, B., Han, S., Wang, M., Yu, Y., Ma, L., Chu, X., Tan, Q., Zhao, Q., Wu, B.(2023) Protein Cell 14: 382-386
- PubMed: 37155313
- DOI: https://doi.org/10.1093/procel/pwac025
- Primary Citation of Related Structures:
7XXH, 7XXI - CAS Key Laboratory of Receptor Research, State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China.
Organizational Affiliation: