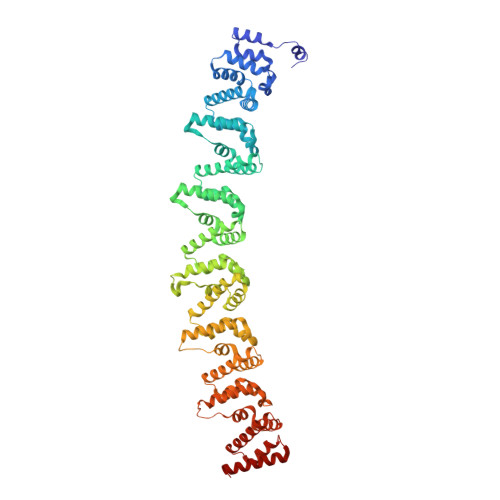
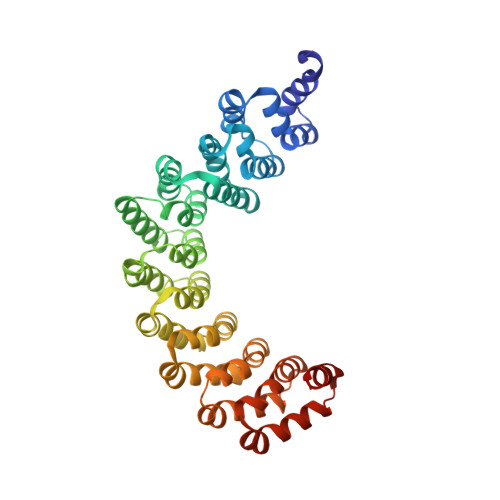Pathogen protein modularity enables elaborate mimicry of a host phosphatase.
Li, H., Wang, J., Kuan, T.A., Tang, B., Feng, L., Wang, J., Cheng, Z., Sklenar, J., Derbyshire, P., Hulin, M., Li, Y., Zhai, Y., Hou, Y., Menke, F.L.H., Wang, Y., Ma, W.(2023) Cell 186: 3196-3207.e17
- PubMed: 37369204
- DOI: https://doi.org/10.1016/j.cell.2023.05.049
- Primary Citation of Related Structures:
7XVI, 7XVK - PubMed Abstract:
Pathogens produce diverse effector proteins to manipulate host cellular processes. However, how functional diversity is generated in an effector repertoire is poorly understood. Many effectors in the devastating plant pathogen Phytophthora contain tandem repeats of the "(L)WY" motif, which are structurally conserved but variable in sequences. Here, we discovered a functional module formed by a specific (L)WY-LWY combination in multiple Phytophthora effectors, which efficiently recruits the serine/threonine protein phosphatase 2A (PP2A) core enzyme in plant hosts. Crystal structure of an effector-PP2A complex shows that the (L)WY-LWY module enables hijacking of the host PP2A core enzyme to form functional holoenzymes. While sharing the PP2A-interacting module at the amino terminus, these effectors possess divergent C-terminal LWY units and regulate distinct sets of phosphoproteins in the host. Our results highlight the appropriation of an essential host phosphatase through molecular mimicry by pathogens and diversification promoted by protein modularity in an effector repertoire.
- The Sainsbury Laboratory, University of East Anglia, Norwich Research Park, Norwich NR4 7UH, UK.
Organizational Affiliation: