Structural basis of lysophosphatidylserine receptor GPR174 ligand recognition and activation.
Liang, J., Inoue, A., Ikuta, T., Xia, R., Wang, N., Kawakami, K., Xu, Z., Qian, Y., Zhu, X., Zhang, A., Guo, C., Huang, Z., He, Y.(2023) Nat Commun 14: 1012-1012
- PubMed: 36823105
- DOI: https://doi.org/10.1038/s41467-023-36575-0
- Primary Citation of Related Structures:
7XV3 - PubMed Abstract:
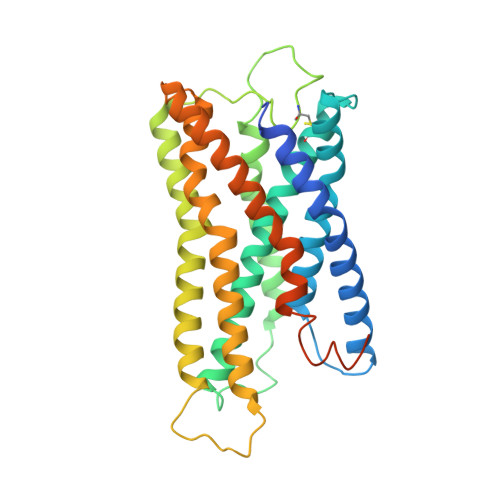
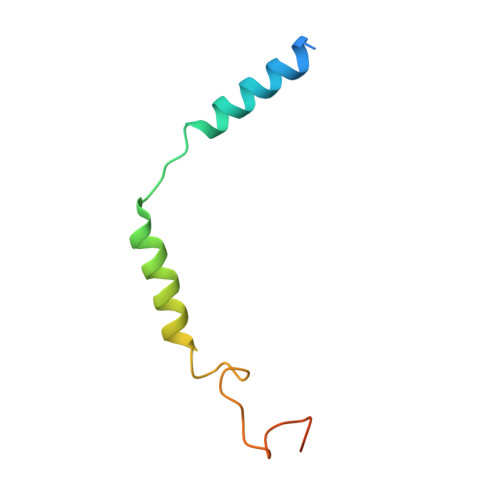
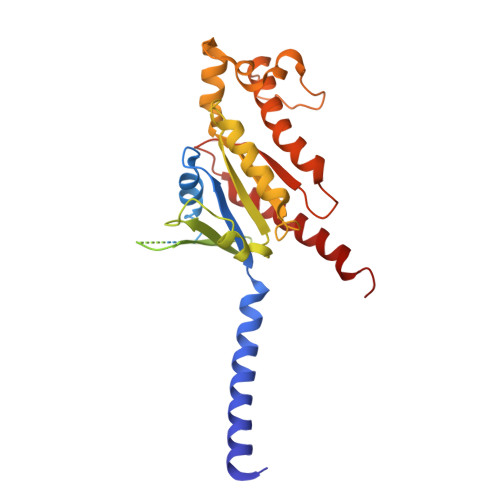
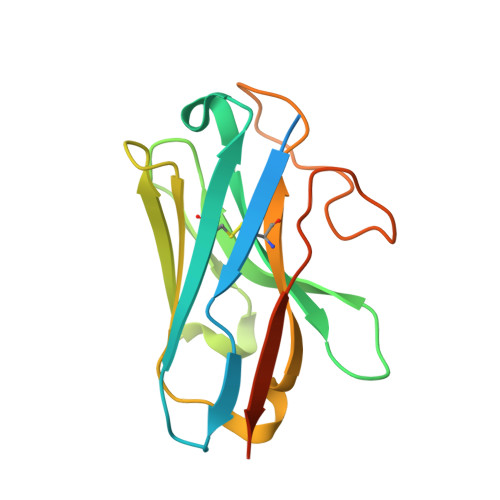
Lysophosphatidylserine (LysoPS) is a lipid mediator that induces multiple cellular responses through binding to GPR174. Here, we present the cryo-electron microscopy (cryo-EM) structure of LysoPS-bound human GPR174 in complex with G s protein. The structure reveals a ligand recognition mode, including the negatively charged head group of LysoPS forms extensive polar interactions with surrounding key residues of the ligand binding pocket, and the L-serine moiety buries deeply into a positive charged cavity in the pocket. In addition, the structure unveils a partially open pocket on transmembrane domain helix (TM) 4 and 5 for a lateral entry of ligand. Finally, the structure reveals a G s engaging mode featured by a deep insertion of a helix 5 (αH5) and extensive polar interactions between receptor and αH5. Taken together, the information revealed by our structural study provides a framework for understanding LysoPS signaling and a rational basis for designing LysoPS receptor-targeting drugs.
- Laboratory of Receptor Structure and Signaling, HIT Center for Life Sciences, School of Life Science and Technology, Harbin Institute of Technology, 150001, Harbin, China.
Organizational Affiliation: