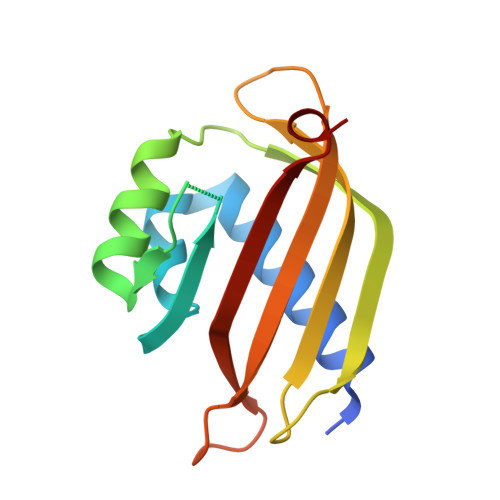
Yin and yang regulation of stress granules by Caprin-1.
Song, D., Kuang, L., Yang, L., Wang, L., Li, H., Li, X., Zhu, Z., Shi, C., Zhu, H., Gong, W.(2022) Proc Natl Acad Sci U S A 119: e2207975119-e2207975119
- PubMed: 36279435
- DOI: https://doi.org/10.1073/pnas.2207975119
- Primary Citation of Related Structures:
7XHF, 7XHG - PubMed Abstract:
Stress granules (SGs) are cytoplasmic biomolecular condensates containing proteins and RNAs in response to stress. Ras-GTPase-activating protein binding protein 1 (G3BP1) is a core SG protein. Caprin-1 and ubiquitin specific peptidase 10 (USP10) interact with G3BP1, facilitating and suppressing SG formation, respectively. The crystal structures of the nuclear transport factor 2-like (NTF2L) domain of G3BP1 in complex with the G3BP1-interacting motif (GIM) of Caprin-1 and USP10 show that both GIMs bind to the same hydrophobic pocket of G3BP1. Moreover, both GIMs suppressed the liquid-liquid phase separation (LLPS) of G3BP1, suggesting that Caprin-1 likely facilitates SG formation via other mechanisms. Thus, we dissected various domains of Caprin-1 and investigated their role in LLPS in vitro and SG formation in cells. The C-terminal domain of Caprin-1 underwent spontaneous LLPS, whereas the N-terminal domain and GIM of Caprin-1 suppressed LLPS of G3BP1. The opposing effect of the N- and C-terminal domains of Caprin-1 on SG formation were demonstrated in cells with or without the endogenous Caprin-1. We propose that the N- and C-terminal domains of Caprin-1 regulate SG formation in a "yin and yang" fashion, mediating the dynamic and reversible assembly of SGs.
- School of Life Sciences, University of Science and Technology of China, Hefei, 230027 China.
Organizational Affiliation: