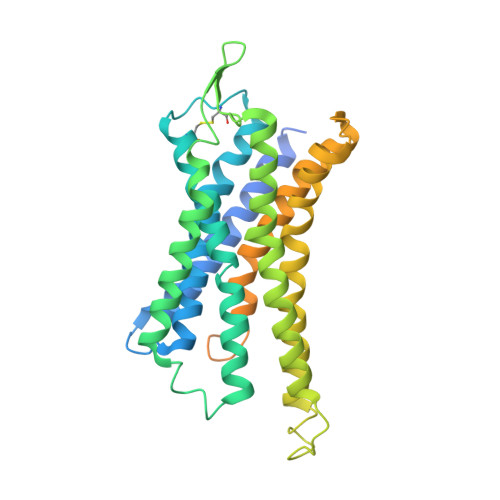
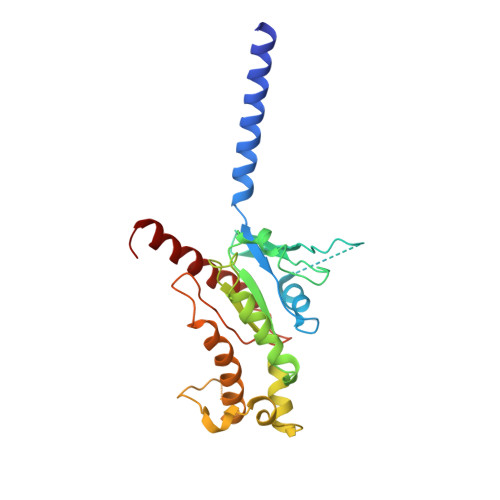
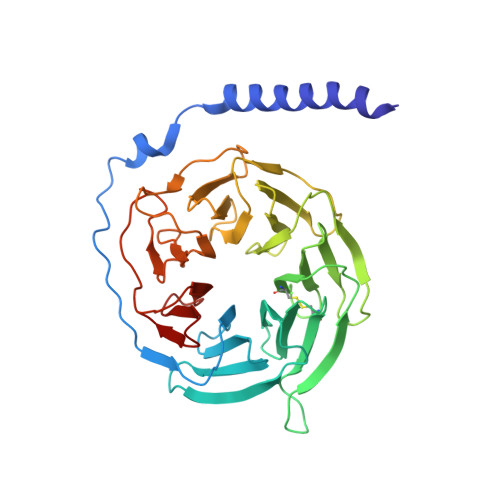
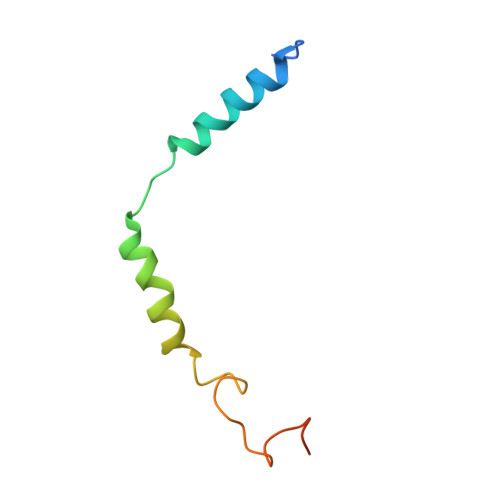
Structure of the human galanin receptor 2 bound to galanin and Gq reveals the basis of ligand specificity and how binding affects the G-protein interface.
Heo, Y., Ishimoto, N., Jeon, Y.E., Yun, J.H., Ohki, M., Anraku, Y., Sasaki, M., Kita, S., Fukuhara, H., Ikuta, T., Kawakami, K., Inoue, A., Maenaka, K., Tame, J.R.H., Lee, W., Park, S.Y.(2022) PLoS Biol 20: e3001714-e3001714
- PubMed: 35913979
- DOI: https://doi.org/10.1371/journal.pbio.3001714
- Primary Citation of Related Structures:
7XBD - PubMed Abstract:
Galanin is a neuropeptide expressed in the central and peripheral nervous systems, where it regulates various processes including neuroendocrine release, cognition, and nerve regeneration. Three G-protein coupled receptors (GPCRs) for galanin have been discovered, which is the focus of efforts to treat diseases including Alzheimer's disease, anxiety, and addiction. To understand the basis of the ligand preferences of the receptors and to assist structure-based drug design, we used cryo-electron microscopy (cryo-EM) to solve the molecular structure of GALR2 bound to galanin and a cognate heterotrimeric G-protein, providing a molecular view of the neuropeptide binding site. Mutant proteins were assayed to help reveal the basis of ligand specificity, and structural comparison between the activated GALR2 and inactive hβ2AR was used to relate galanin binding to the movements of transmembrane (TM) helices and the G-protein interface.
- Structural Biochemistry & Molecular Biophysics Laboratory, Department of Biochemistry, College of Life Science and Biotechnology, Yonsei University, Seoul, Korea.
Organizational Affiliation: