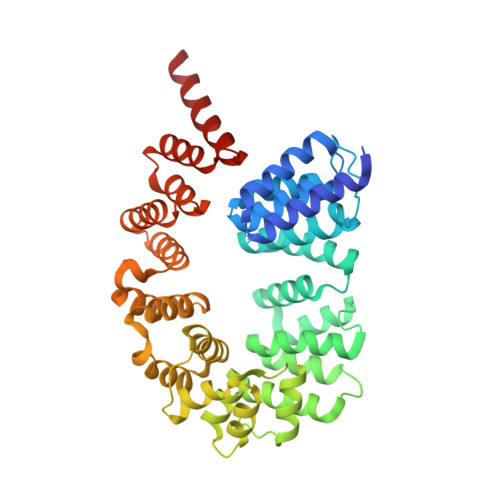
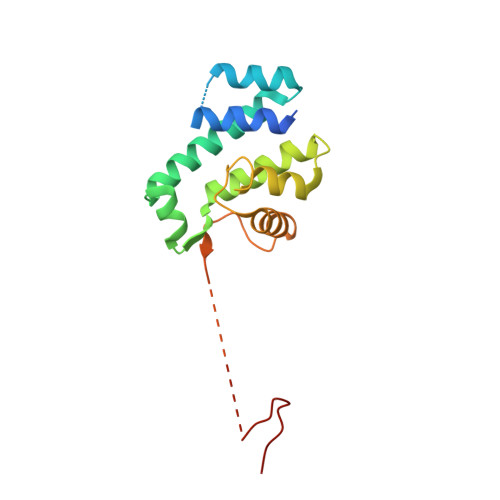
Structural basis of flagellar motility regulation by the MogR repressor and the GmaR antirepressor in Listeria monocytogenes.
Cho, S.Y., Na, H.W., Oh, H.B., Kwak, Y.M., Song, W.S., Park, S.C., Jeon, W.J., Cho, H., Oh, B.C., Park, J., Kang, S.G., Lee, G.S., Yoon, S.I.(2022) Nucleic Acids Res 50: 11315-11330
- PubMed: 36283692
- DOI: https://doi.org/10.1093/nar/gkac815
- Primary Citation of Related Structures:
7X9R, 7X9S - PubMed Abstract:
The pathogenic Listeria monocytogenes bacterium produces the flagellum as a locomotive organelle at or below 30°C outside the host, but it halts flagellar expression at 37°C inside the human host to evade the flagellum-induced immune response. Listeria monocytogenes GmaR is a thermosensor protein that coordinates flagellar expression by binding the master transcriptional repressor of flagellar genes (MogR) in a temperature-responsive manner. To understand the regulatory mechanism whereby GmaR exerts the antirepression activity on flagellar expression, we performed structural and mutational analyses of the GmaR-MogR system. At or below 30°C, GmaR exists as a functional monomer and forms a circularly enclosed multidomain structure via an interdomain interaction. GmaR in this conformation recognizes MogR using the C-terminal antirepressor domain in a unique dual binding mode and mediates the antirepressor function through direct competition and spatial restraint mechanisms. Surprisingly, at 37°C, GmaR rapidly forms autologous aggregates that are deficient in MogR neutralization capabilities.
- Division of Biomedical Convergence, College of Biomedical Science, Kangwon National University, Chuncheon 24341, Republic of Korea.
Organizational Affiliation: