Structural basis of transcription regulation by CNC family transcription factor, Nrf2.
Sengoku, T., Shiina, M., Suzuki, K., Hamada, K., Sato, K., Uchiyama, A., Kobayashi, S., Oguni, A., Itaya, H., Kasahara, K., Moriwaki, H., Watanabe, C., Honma, T., Okada, C., Baba, S., Ohta, T., Motohashi, H., Yamamoto, M., Ogata, K.(2022) Nucleic Acids Res 50: 12543-12557
- PubMed: 36454022
- DOI: https://doi.org/10.1093/nar/gkac1102
- Primary Citation of Related Structures:
7X5E, 7X5F, 7X5G - PubMed Abstract:
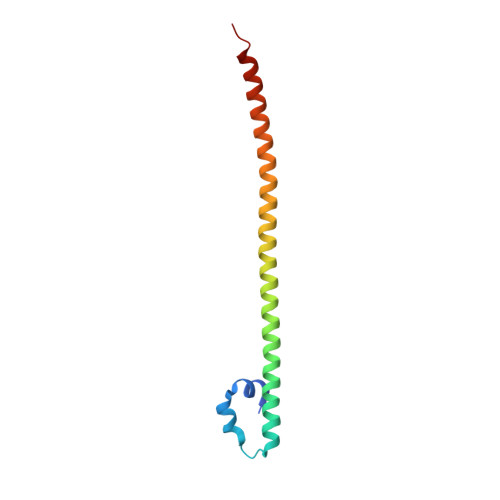
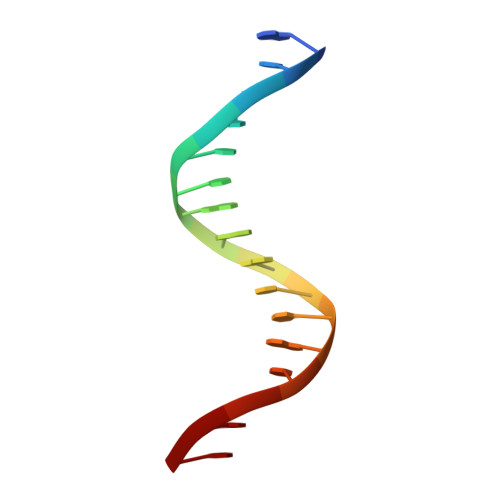
Several basic leucine zipper (bZIP) transcription factors have accessory motifs in their DNA-binding domains, such as the CNC motif of CNC family or the EHR motif of small Maf (sMaf) proteins. CNC family proteins heterodimerize with sMaf proteins to recognize CNC-sMaf binding DNA elements (CsMBEs) in competition with sMaf homodimers, but the functional role of the CNC motif remains elusive. In this study, we report the crystal structures of Nrf2/NFE2L2, a CNC family protein regulating anti-stress transcriptional responses, in a complex with MafG and CsMBE. The CNC motif restricts the conformations of crucial Arg residues in the basic region, which form extensive contact with the DNA backbone phosphates. Accordingly, the Nrf2-MafG heterodimer has approximately a 200-fold stronger affinity for CsMBE than canonical bZIP proteins, such as AP-1 proteins. The high DNA affinity of the CNC-sMaf heterodimer may allow it to compete with the sMaf homodimer on target genes without being perturbed by other low-affinity bZIP proteins with similar sequence specificity.
- Department of Biochemistry, Yokohama City University Graduate School of Medicine, Yokohama 236-0004, Japan.
Organizational Affiliation: