Cryo-electron microscopy structure of the H3-H4 octasome: A nucleosome-like particle without histones H2A and H2B.
Nozawa, K., Takizawa, Y., Pierrakeas, L., Sogawa-Fujiwara, C., Saikusa, K., Akashi, S., Luk, E., Kurumizaka, H.(2022) Proc Natl Acad Sci U S A 119: e2206542119-e2206542119
- PubMed: 36322721
- DOI: https://doi.org/10.1073/pnas.2206542119
- Primary Citation of Related Structures:
7X57, 7X58, 7YOZ - PubMed Abstract:
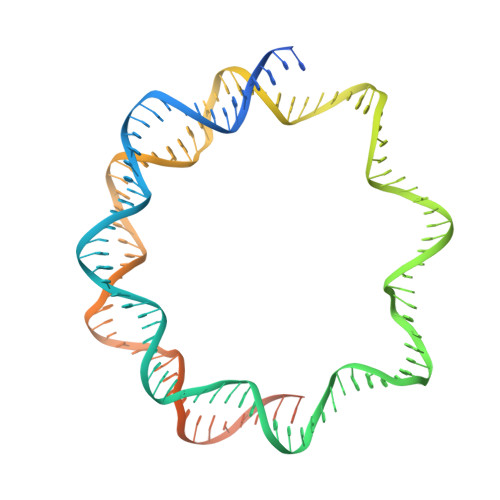
The canonical nucleosome, which represents the major packaging unit of eukaryotic chromatin, has an octameric core composed of two histone H2A-H2B and H3-H4 dimers with ∼147 base pairs (bp) of DNA wrapped around it. Non-nucleosomal particles with alternative histone stoichiometries and DNA wrapping configurations have been found, and they could profoundly influence genome architecture and function. Using cryo-electron microscopy, we solved the structure of the H3-H4 octasome, a nucleosome-like particle with a di-tetrameric core consisting exclusively of the H3 and H4 histones. The core is wrapped by ∼120 bp of DNA in 1.5 negative superhelical turns, forming two stacked disks that are connected by a H4-H4' four-helix bundle. Three conformations corresponding to alternative interdisk angles were observed, indicating the flexibility of the H3-H4 octasome structure. In vivo crosslinking experiments detected histone-histone interactions consistent with the H3-H4 octasome model, suggesting that H3-H4 octasomes or related structural features exist in cells.
- Laboratory of Chromatin Structure and Function, Institute for Quantitative Biosciences, The University of Tokyo, Tokyo 113-0032, Japan.
Organizational Affiliation: