Structure of a human NHE3-CHP1 complex in the autoinhibited state
Dong, Y., Li, H., Ilie, A., Gao, Y., Boucher, A., Zhang, X.C., Orlowski, J., Zhao, Y.(2022) Sci Adv
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2022) Sci Adv
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Sodium/hydrogen exchanger 3 | 626 | Homo sapiens | Mutation(s): 0 Gene Names: SLC9A3, NHE3 Membrane Entity: Yes | 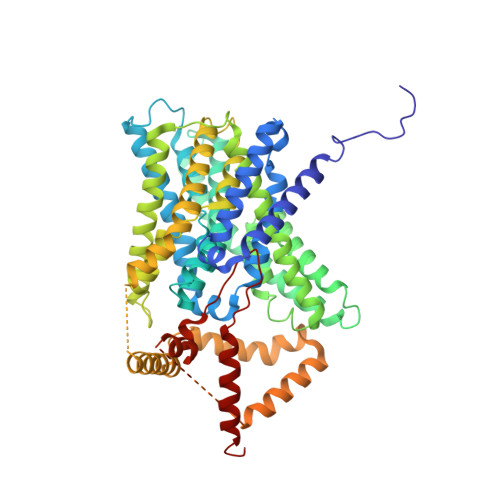 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P48764 (Homo sapiens) Explore P48764 Go to UniProtKB: P48764 | |||||
PHAROS: P48764 GTEx: ENSG00000066230 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P48764 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Calcineurin B homologous protein 1 | 185 | Homo sapiens | Mutation(s): 0 Gene Names: CHP1, CHP Membrane Entity: Yes | 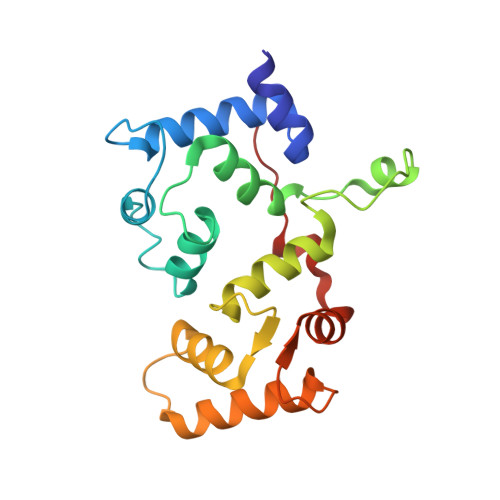 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q99653 (Homo sapiens) Explore Q99653 Go to UniProtKB: Q99653 | |||||
PHAROS: Q99653 GTEx: ENSG00000187446 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q99653 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 85R (Subject of Investigation/LOI) Query on 85R | E [auth A], L [auth B] | [(2~{R})-2-hexadecanoyloxy-3-[oxidanyl-[(2~{S},3~{S},5~{R},6~{S})-2,3,4,5,6-pentakis(oxidanyl)cyclohexyl]oxy-phosphoryl]oxy-propyl] hexadecanoate C41 H79 O13 P IBUKXRINTKQBRQ-PLWKSVIPSA-N |  | ||
| PGT Query on PGT | F [auth A] G [auth A] H [auth A] I [auth A] J [auth A] | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE C40 H79 O10 P KBPVYRBBONZJHF-AMAPPZPBSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Chinese Academy of Sciences | China | XDB37030304 |