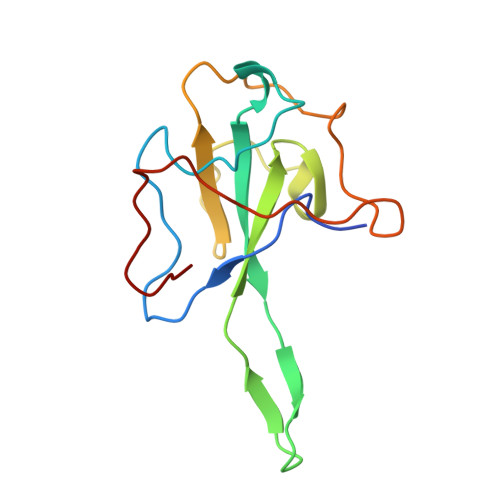
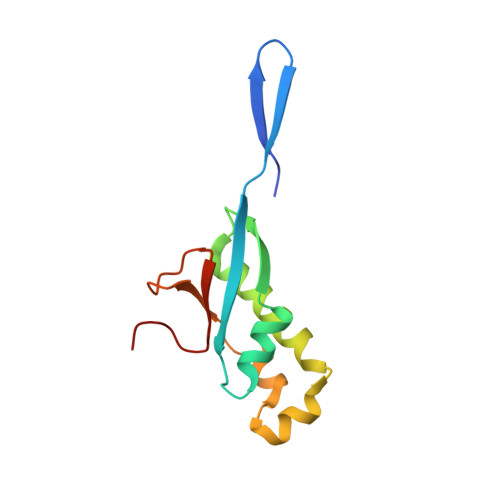
Structural insights into ribonucleoprotein dissociation by nucleocapsid protein interacting with non-structural protein 3 in SARS-CoV-2.
Ni, X., Han, Y., Zhou, R., Zhou, Y., Lei, J.(2023) Commun Biol 6: 193-193
- PubMed: 36806252
- DOI: https://doi.org/10.1038/s42003-023-04570-2
- Primary Citation of Related Structures:
7VNU, 7WZO - PubMed Abstract:
The coronavirus nucleocapsid (N) protein interacts with non-structural protein 3 (Nsp3) to facilitate viral RNA synthesis and stabilization. However, structural information on the N-Nsp3 complex is limited. Here, we report a 2.6 Å crystal structure of the N-terminal domain (NTD) of the N protein in complex with the ubiquitin-like domain 1 (Ubl1) of Nsp3 in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). One NTD and two Ubl1s formed a stable heterotrimer. We performed mutational analysis to reveal the key residues for this interaction. We confirmed the colocalization of SARS-CoV-2 N and Nsp3 in Huh-7 cells. N-Ubl1 interaction also exists in SARS-CoV and Middle East respiratory syndrome coronavirus. We found that SARS-CoV-2 Ubl1 competes with RNA to bind N protein in a dose-dependent manner. Based on our results, we propose a model for viral ribonucleoprotein dissociation through N protein binding to Ubl1 of Nsp3.
- National Clinical Research Center for Geriatrics, West China Hospital, Sichuan University, Chengdu, Sichuan, China.
Organizational Affiliation: