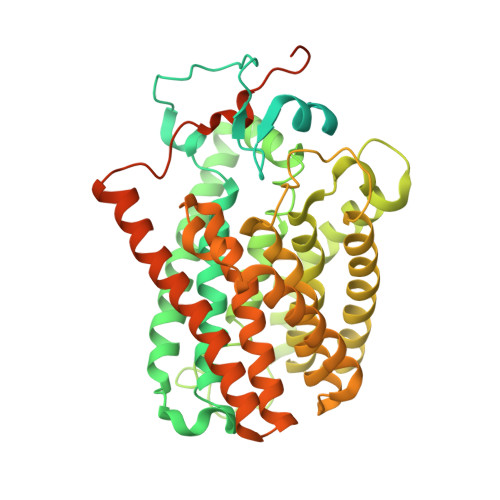
Structure of the Arabidopsis guard cell anion channel SLAC1 suggests activation mechanism by phosphorylation.
Li, Y., Ding, Y., Qu, L., Li, X., Lai, Q., Zhao, P., Gao, Y., Xiang, C., Cang, C., Liu, X., Sun, L.(2022) Nat Commun 13: 2511-2511
- PubMed: 35523967
- DOI: https://doi.org/10.1038/s41467-022-30253-3
- Primary Citation of Related Structures:
7WNQ - PubMed Abstract:
Stomata play a critical role in the regulation of gas exchange and photosynthesis in plants. Stomatal closure participates in multiple stress responses, and is regulated by a complex network including abscisic acid (ABA) signaling and ion-flux-induced turgor changes. The slow-type anion channel SLAC1 has been identified to be a central controller of stomatal closure and phosphoactivated by several kinases. Here, we report the structure of SLAC1 in Arabidopsis thaliana (AtSLAC1) in an inactivated, closed state. The cytosolic amino (N)-terminus and carboxyl (C)-terminus of AtSLAC1 are partially resolved and form a plug-like structure which packs against the transmembrane domain (TMD). Breaking the interactions between the cytosolic plug and transmembrane domain triggers channel activation. An inhibition-release model is proposed for SLAC1 activation by phosphorylation that the cytosolic plug dissociates from the transmembrane domain upon phosphorylation, and induces conformational changes to open the pore. These findings facilitate our understanding of the regulation of SLAC1 activity and stomatal aperture in plants.
- Department of Neurology, the First Affiliated Hospital of USTC, MOE Key Laboratory for Membraneless Organelles and Cellular Dynamics, Hefei National Laboratory for Physical Sciences at the Microscale, CAS Centre for Excellence in Molecular Cell Science, Biomedical Sciences and Health Laboratory of Anhui Province, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, 230027, China.
Organizational Affiliation: