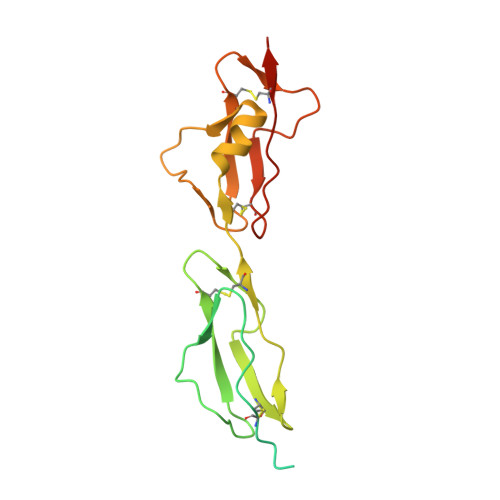
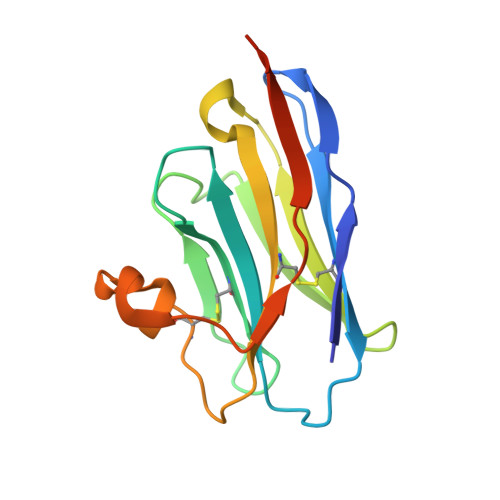
Antibody recognition of complement factor H reveals a flexible loop involved in atypical hemolytic uremic syndrome pathogenesis.
Yokoo, T., Tanabe, A., Yoshida, Y., Caaveiro, J.M.M., Nakakido, M., Ikeda, Y., Fujimura, Y., Matsumoto, M., Entzminger, K., Maruyama, T., Okumura, C.J., Nangaku, M., Tsumoto, K.(2022) J Biological Chem 298: 101962-101962
- PubMed: 35452676
- DOI: https://doi.org/10.1016/j.jbc.2022.101962
- Primary Citation of Related Structures:
7WKI - PubMed Abstract:
Atypical hemolytic uremic syndrome (aHUS) is a disease associated with dysregulation of the immune complement system, especially of the alternative pathway (AP). Complement factor H (CFH), consisting of 20 domains called complement control protein (CCP1-20), downregulates the AP as a cofactor for mediating C3 inactivation by complement factor I. However, anomalies related to CFH are known to cause excessive complement activation and cytotoxicity. In aHUS, mutations and the presence of anti-CFH autoantibodies (AAbs) have been reported as plausible causes of CFH dysfunction, and it is known that CFH-related aHUS carries a high probability of end-stage renal disease. Elucidating the detailed functions of CFH at the molecular level will help to understand aHUS pathogenesis. Herein, we used biophysical data to reveal that a heavy-chain antibody fragment, termed VHH4, recognized CFH with high affinity. Hemolytic assays also indicated that VHH4 disrupted the protective function of CFH on sheep erythrocytes. Furthermore, X-ray crystallography revealed that VHH4 recognized the Leu1181-Leu1189 CCP20 loop, a known anti-CFH AAbs epitope. We next analyzed the dynamics of the C-terminal region of CFH and showed that the epitopes recognized by anti-CFH AAbs and VHH4 were the most flexible regions in CCP18-20. Finally, we conducted mutation analyses to elucidate the mechanism of VHH4 recognition of CFH and revealed that VHH4 inserts the Trp1183 CCP20 residue of CFH into the pocket formed by the complementary determining region 3 loop. These results suggested that anti-CFH AAbs may adopt a similar molecular mechanism to recognize the flexible loop of Leu1181-Leu1189 CCP20 , leading to aHUS pathogenesis.
- Department of Chemistry and Biotechnology, School of Engineering, The University of Tokyo, Bunkyo-ku, Tokyo, Japan.
Organizational Affiliation: