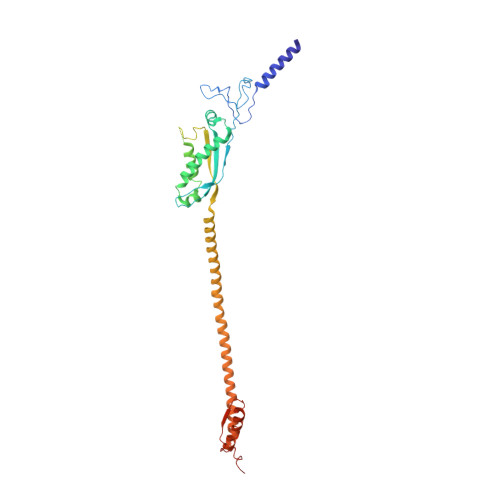
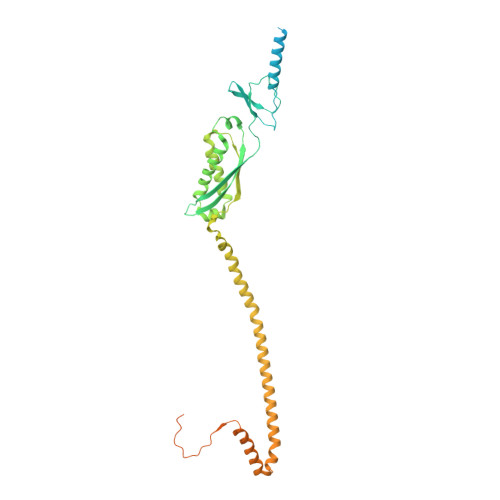
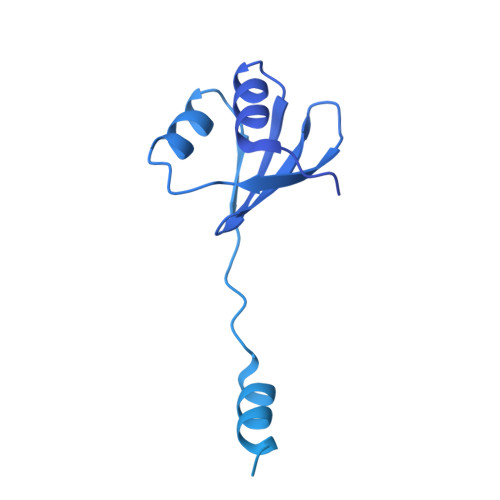
Cryo-EM structure of the entire FtsH-HflKC AAA protease complex.
Qiao, Z., Yokoyama, T., Yan, X.F., Beh, I.T., Shi, J., Basak, S., Akiyama, Y., Gao, Y.G.(2022) Cell Rep 39: 110890-110890
- PubMed: 35649372
- DOI: https://doi.org/10.1016/j.celrep.2022.110890
- Primary Citation of Related Structures:
7WI3, 7WI4 - PubMed Abstract:
The membrane-bound AAA protease FtsH is the key player controlling protein quality in bacteria. Two single-pass membrane proteins, HflK and HflC, interact with FtsH to modulate its proteolytic activity. Here, we present structure of the entire FtsH-HflKC complex, comprising 12 copies of both HflK and HflC, all of which interact reciprocally to form a cage, as well as four FtsH hexamers with periplasmic domains and transmembrane helices enclosed inside the cage and cytoplasmic domains situated at the base of the cage. FtsH K61/D62/S63 in the β2-β3 loop in the periplasmic domain directly interact with HflK, contributing to complex formation. Pull-down and in vivo enzymatic activity assays validate the importance of the interacting interface for FtsH-HflKC complex formation. Structural comparison with the substrate-bound human m-AAA protease AFG3L2 offers implications for the HflKC cage in modulating substrate access to FtsH. Together, our findings provide a better understanding of FtsH-type AAA protease holoenzyme assembly and regulation.
- School of Biological Sciences, Nanyang Technological University, Singapore 637551, Singapore; NTU Institute of Structural Biology, Nanyang Technological University, Singapore 639798, Singapore.
Organizational Affiliation: