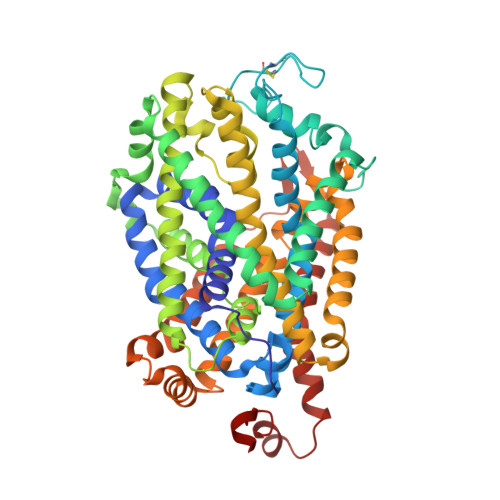
Structural insights into GABA transport inhibition using an engineered neurotransmitter transporter.
Joseph, D., Nayak, S.R., Penmatsa, A.(2022) EMBO J 41: e110735-e110735
- PubMed: 35796008
- DOI: https://doi.org/10.15252/embj.2022110735
- Primary Citation of Related Structures:
7WGD, 7WGT, 7WLW - PubMed Abstract:
γ-aminobutyric acid (GABA) is the major inhibitory neurotransmitter, and its levels in the synaptic space are controlled by the GABA transporter isoforms (GATs). GATs are structurally related to biogenic amine transporters but display interactions with distinct inhibitors used as anti-epileptics. In this study, we engineer the binding pocket of Drosophila melanogaster dopamine transporter to resemble GAT1 and determine high-resolution X-ray structures of the modified transporter in the substrate-free state and in complex with GAT1 inhibitors NO711 and SKF89976a that are analogs of tiagabine, a medication prescribed for the treatment of partial seizures. We observe that the primary binding site undergoes substantial shifts in subsite architecture in the modified transporter to accommodate the two GAT1 inhibitors. We also observe that SKF89976a additionally interacts at an allosteric site in the extracellular vestibule, yielding an occluded conformation. Interchanging SKF89976a interacting residue in the extracellular loop 4 between GAT1 and dDAT suggests a role for this motif in the selective control of neurotransmitter uptake. Our findings, therefore, provide vital insights into the organizational principles dictating GAT1 activity and inhibition.
- Molecular Biophysics Unit, Indian Institute of Science, Bangalore, India.
Organizational Affiliation: