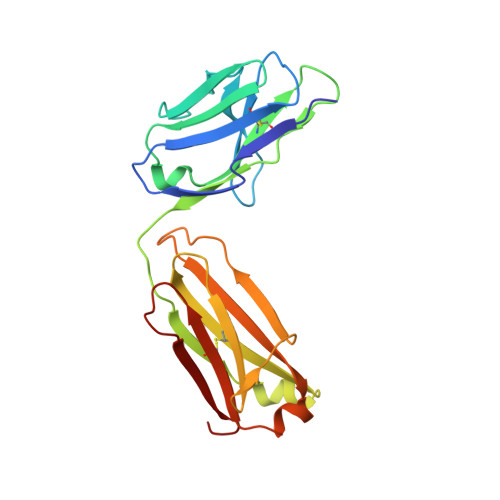
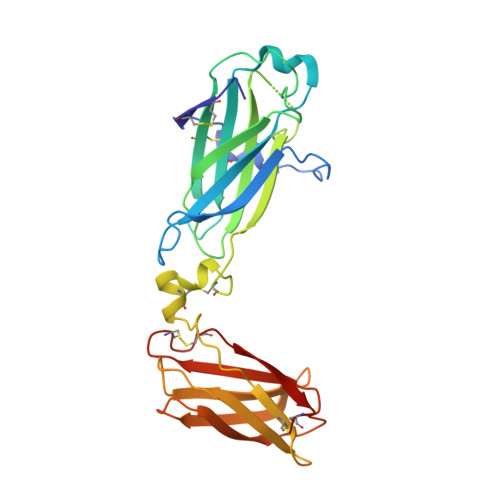
Structural basis of interleukin-17B receptor in complex with a neutralizing antibody for guiding humanization and affinity maturation.
Lee, W.H., Chen, X., Liu, I.J., Lee, J.H., Hu, C.M., Wu, H.C., Wang, S.K., Lee, W.H., Ma, C.(2022) Cell Rep 41: 111555-111555
- PubMed: 36288706
- DOI: https://doi.org/10.1016/j.celrep.2022.111555
- Primary Citation of Related Structures:
7WG3 - PubMed Abstract:
Upregulation of interleukin-17 receptor B (IL-17RB) is known to be oncogenic, while other IL-17 receptors and ligands are generally involved in pro-inflammatory pathways. We identify a mouse neutralizing monoclonal antibody (mAb) D9, which blocks the IL-17RB/IL-17B pathway and inhibits pancreatic tumorigenesis in an orthotopic mouse model. The X-ray crystal structure of the IL-17RB ectodomain in complex with its neutralizing antibody D9 shows that D9 binds to a predicted ligand binding interface and engages with the A'-A loop of IL-17RB fibronectin III domain 1 in a unique conformational state. This structure also provides important paratope information to guide the design of antibody humanization and affinity maturation of D9, resulting in a humanized 1B12 antibody with marginal affinity loss and effective neutralization of IL-17B/IL-17RB signaling to impede tumorigenesis in a mouse xenograft model.
- Genomics Research Center, Academia Sinica, Taipei, Taiwan; Chemical Biology and Molecular Biophysics, Taiwan International Graduate Program, Academia Sinica, Taipei, Taiwan; Department of Chemistry, National Tsing Hua University, Hsinchu, Taiwan.
Organizational Affiliation: