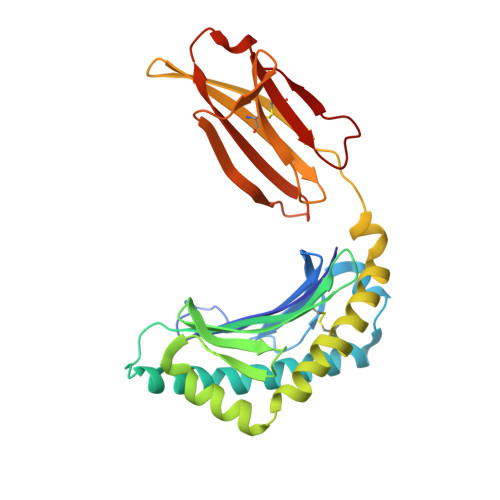
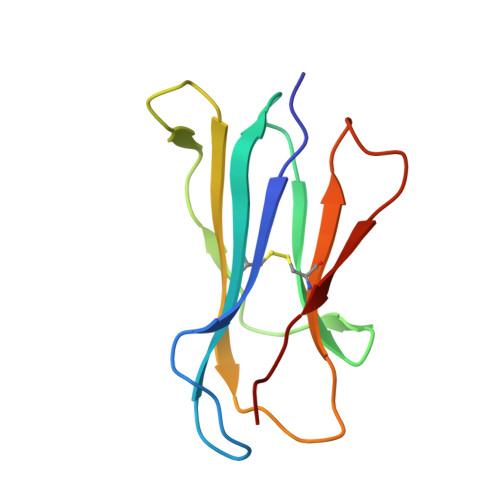
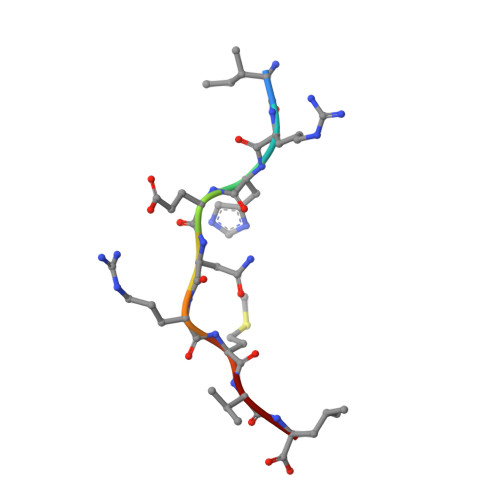
A Wider and Deeper Peptide-Binding Groove for the Class I Molecules from B15 Compared with B19 Chickens Correlates with Relative Resistance to Marek's Disease.
Han, L., Wu, S., Zhang, T., Peng, W., Zhao, M., Yue, C., Wen, W., Cai, W., Li, M., Wallny, H.J., Avila, D.W., Mwangi, W., Nair, V., Ternette, N., Guo, Y., Zhao, Y., Chai, Y., Qi, J., Liang, H., Gao, G.F., Kaufman, J., Liu, W.J.(2023) J Immunol 210: 668-680
- PubMed: 36695776
- DOI: https://doi.org/10.4049/jimmunol.2200211
- Primary Citation of Related Structures:
7WBG, 7WBI - PubMed Abstract:
The chicken MHC is known to confer decisive resistance or susceptibility to various economically important pathogens, including the iconic oncogenic herpesvirus that causes Marek's disease (MD). Only one classical class I gene, BF2, is expressed at a high level in chickens, so it was relatively easy to discern a hierarchy from well-expressed thermostable fastidious specialist alleles to promiscuous generalist alleles that are less stable and expressed less on the cell surface. The class I molecule BF2*1901 is better expressed and more thermostable than the closely related BF2*1501, but the peptide motif was not simpler as expected. In this study, we confirm for newly developed chicken lines that the chicken MHC haplotype B15 confers resistance to MD compared with B19. Using gas phase sequencing and immunopeptidomics, we find that BF2*1901 binds a greater variety of amino acids in some anchor positions than does BF2*1501. However, by x-ray crystallography, we find that the peptide-binding groove of BF2*1901 is narrower and shallower. Although the self-peptides that bound to BF2*1901 may appear more various than those of BF2*1501, the structures show that the wider and deeper peptide-binding groove of BF2*1501 allows stronger binding and thus more peptides overall, correlating with the expected hierarchies for expression level, thermostability, and MD resistance. Our study provides a reasonable explanation for greater promiscuity for BF2*1501 compared with BF2*1901, corresponding to the difference in resistance to MD.
- State Key Laboratory of Veterinary Biotechnology, Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Harbin, China.
Organizational Affiliation: