Structural basis for the toxin-coregulated pilus-dependent secretion of Vibrio cholerae colonization factor.
Oki, H., Kawahara, K., Iimori, M., Imoto, Y., Nishiumi, H., Maruno, T., Uchiyama, S., Muroga, Y., Yoshida, A., Yoshida, T., Ohkubo, T., Matsuda, S., Iida, T., Nakamura, S.(2022) Sci Adv 8: eabo3013-eabo3013
- PubMed: 36240278
- DOI: https://doi.org/10.1126/sciadv.abo3013
- Primary Citation of Related Structures:
7W63, 7W64, 7W65 - PubMed Abstract:
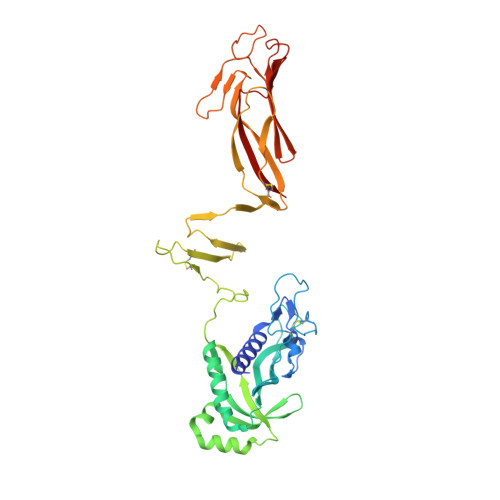
Colonization of the host intestine is the most important step in Vibrio cholerae infection. The toxin-coregulated pilus (TCP), an operon-encoded type IVb pilus (T4bP), plays a crucial role in this process, which requires an additional secreted protein, TcpF, encoded on the same TCP operon; however, its mechanisms of secretion and function remain elusive. Here, we demonstrated that TcpF interacts with the minor pilin, TcpB, of TCP and elucidated the crystal structures of TcpB alone and in complex with TcpF. The structural analyses reveal how TCP recognizes TcpF and its secretory mechanism via TcpB-dependent pilus elongation and retraction. Upon binding to TCP, TcpF forms a flower-shaped homotrimer with its flexible N terminus hooked onto the trimeric interface of TcpB. Thus, the interaction between the minor pilin and the N terminus of the secreted protein, namely, the T4bP secretion signal, is key for V. cholerae colonization and is a new potential therapeutic target.
- Department of Infection Metagenomics, Genome Information Research Center, Research Institute for Microbial Diseases, Osaka University, Osaka, Japan.
Organizational Affiliation: