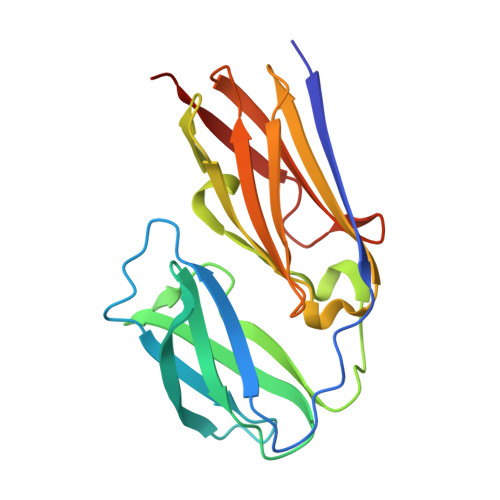
Crystal Structure Analysis and IgE Epitope Mapping of Allergic Predominant Region in Scylla paramamosain Filamin C, Scy p 9.
He, X.R., Yang, Y., Kang, S., Chen, Y.X., Zheng, P.Y., Chen, G.X., Chen, X.M., Cao, M.J., Jin, T., Liu, G.M.(2022) J Agric Food Chem 70: 1282-1292
- PubMed: 35040643
- DOI: https://doi.org/10.1021/acs.jafc.1c07922
- Primary Citation of Related Structures:
7VZO - PubMed Abstract:
Filamin C (FLN c) is a novel allergen in shellfish. In this study, FLN c from Scylla paramamosain was divided into three regions for recombinant expression based on the number of domains and amino acids. Using dot blot and basophil activation tests, the allergic predominant region of FLN c was determined to be 336-531 amino acid positions (named FLN c-M). It was confirmed that by X-ray diffraction, the crystal structure of FLN c-M with immunoglobulin-like folding at a resolution of 1.7 Å was obtained. The monomer was a barrel structure composed of 16 β-strands and 2 α-helices. Three conformational epitopes were predicted, six linear epitopes were verified by serological test, and they were positioned on the crystal structure of FLN c-M. For the first time, the crystal structure of the allergic predominant region of FLN c was determined, and it provided an accurate template for the localization of IgE epitopes.
- College of Marine Food and Biological Engineering, Xiamen Key Laboratory of Marine Functional Food, Fujian Provincial Engineering Technology Research Center of Marine Functional Food, Jimei University, Xiamen, Fujian 361000, China.
Organizational Affiliation: