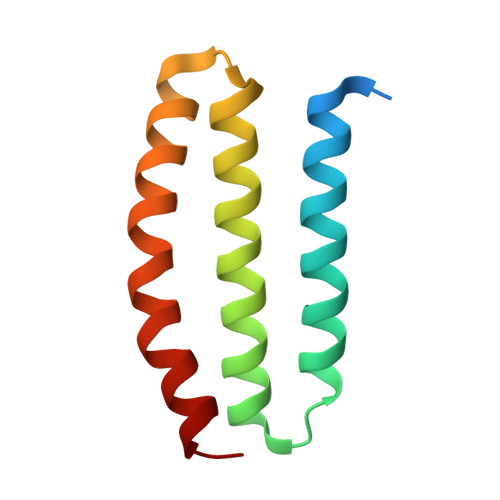
Structural basis of copper binding by a dimeric periplasmic protein forming a six-helical bundle.
Yang, J., Gao, M., Wang, J., He, C., Wang, X., Liu, L.(2022) J Inorg Biochem 229: 111728-111728
- PubMed: 35066349
- DOI: https://doi.org/10.1016/j.jinorgbio.2022.111728
- Primary Citation of Related Structures:
7VW0, 7VW1, 7VW2 - PubMed Abstract:
Bacteria maintain copper balance by various copper response mechanisms. A plasmid gene encoding a methionine rich protein targeted to periplasm is adjacent to the sil operon that confers heavy metal resistance. However, the gene product Orf91 has not been characterized before. Using X-ray crystallography, we solved the structures of Orf91 in apo, cuprous ion-bound, and cupric ion-bound forms. An Orf91 protomer consists of three helices of which the C-terminal two helices belong to domain of unknown function 305 (DUF305), and two Orf91s dimerize into a six-helical bundle. The MxxHH motif specific for DUF305 is critical for cuprous ion binding, and the MxxMxxMHxxMM motif in the N-terminal helix contributes to cupric ion binding. The first histidine of MxxHH shows alternative conformations related to the redox state of copper ion. We suggest that Orf91 is an adaptable copper sponge in the periplasmic space.
- School of Life Sciences, Anhui University, 111 Jiulong Road, Hefei, Anhui 230601, China.
Organizational Affiliation: