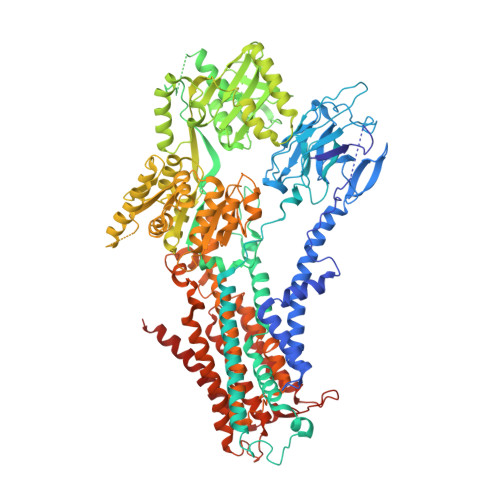
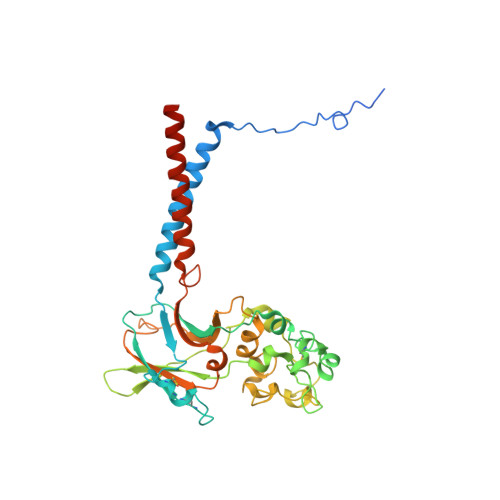
Cryo-EM of the ATP11C flippase reconstituted in Nanodiscs shows a distended phospholipid bilayer inner membrane around transmembrane helix 2.
Nakanishi, H., Hayashida, K., Nishizawa, T., Oshima, A., Abe, K.(2022) J Biological Chem 298: 101498-101498
- PubMed: 34922944
- DOI: https://doi.org/10.1016/j.jbc.2021.101498
- Primary Citation of Related Structures:
7VSG, 7VSH - PubMed Abstract:
ATP11C is a member of the P4-ATPase flippase family that mediates translocation of phosphatidylserine (PtdSer) across the lipid bilayer. In order to characterize the structure and function of ATP11C in a model natural lipid environment, we revisited and optimized a quick procedure for reconstituting ATP11C into Nanodiscs using methyl-β-cyclodextrin as a reagent for the detergent removal. ATP11C was efficiently reconstituted with the endogenous lipid, or the mixture of endogenous lipid and synthetic dioleoylphosphatidylcholine (DOPC)/dioleoylphosphatidylserine (DOPS), all of which retained the ATPase activity. We obtained 3.4 Å and 3.9 Å structures using single-particle cryo-electron microscopy (cryo-EM) of AlF- and BeF-stabilized ATP11C transport intermediates, respectively, in a bilayer containing DOPS. We show that the latter exhibited a distended inner membrane around ATP11C transmembrane helix 2, possibly reflecting the perturbation needed for phospholipid release to the lipid bilayer. Our structures of ATP11C in the lipid membrane indicate that the membrane boundary varies upon conformational changes of the enzyme and is no longer flat around the protein, a change that likely contributes to phospholipid translocation across the membrane leaflets.
- Cellular and Structural Physiology Institute, Nagoya University, Nagoya, Japan.
Organizational Affiliation: