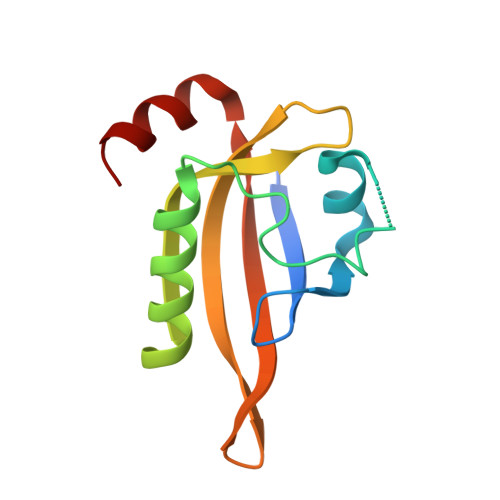
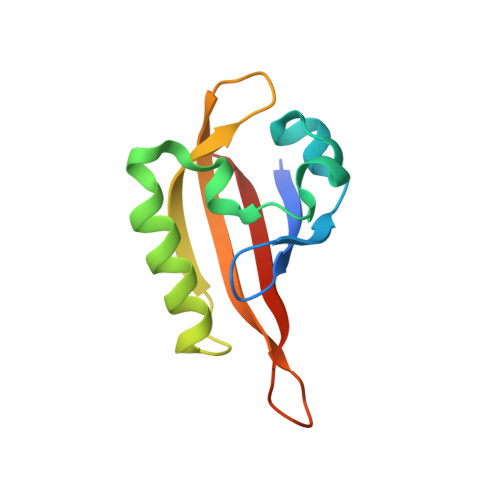
Structural insight into the ligand binding mechanism of aryl hydrocarbon receptor.
Dai, S., Qu, L., Li, J., Zhang, Y., Jiang, L., Wei, H., Guo, M., Chen, X., Chen, Y.(2022) Nat Commun 13: 6234-6234
- PubMed: 36266304
- DOI: https://doi.org/10.1038/s41467-022-33858-w
- Primary Citation of Related Structures:
7VNA, 7VNH, 7VNI - PubMed Abstract:
The aryl hydrocarbon receptor (AHR), a member of the basic helix-loop-helix (bHLH) Per-Arnt-Sim (PAS) family of transcription factors, plays important roles in regulating xenobiotic metabolism, cellular differentiation, stem cell maintenance, as well as immunity. More recently, AHR has gained significant interest as a drug target for the development of novel cancer immunotherapy drugs. Detailed understanding of AHR-ligand binding has been hampered for decades by the lack of a three-dimensional structure of the AHR PAS-B domain. Here, we present multiple crystal structures of the Drosophila AHR PAS-B domain, including its apo, ligand-bound, and AHR nuclear translocator (ARNT) PAS-B-bound forms. Together with biochemical and cellular assays, our data reveal structural features of the AHR PAS-B domain, provide insights into the mechanism of AHR ligand binding, and provide the structural basis for the future development of AHR-targeted therapeutics.
- Department of Oncology, NHC Key Laboratory of Cancer Proteomics & State Local Joint Engineering Laboratory for Anticancer Drugs, National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, Hunan, 410008, China.
Organizational Affiliation: