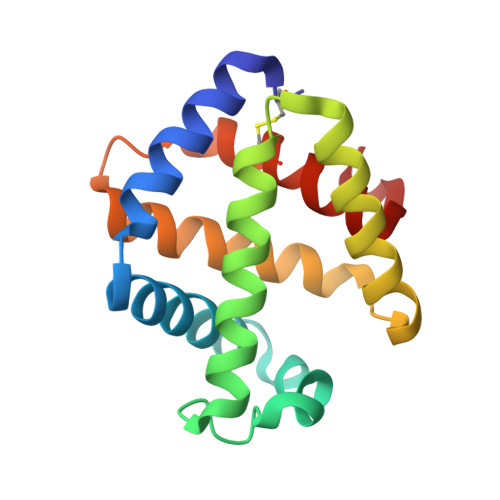
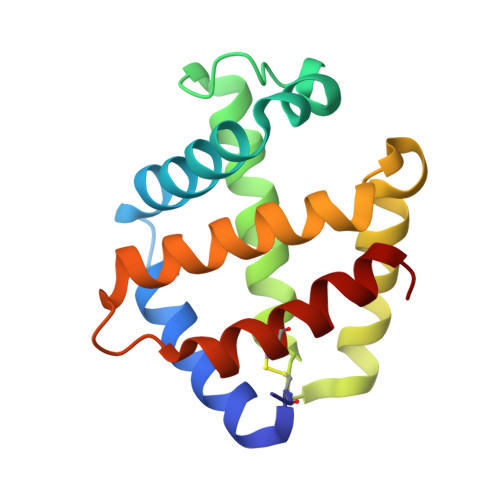
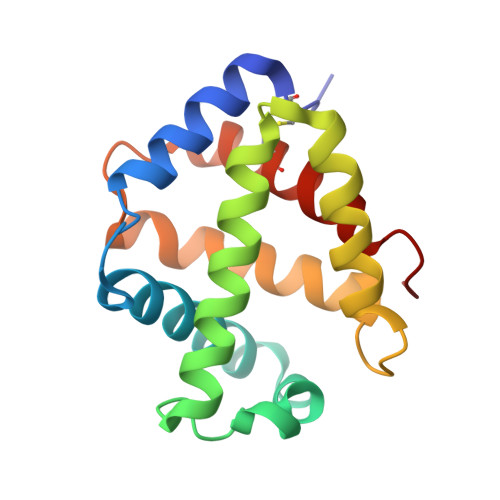
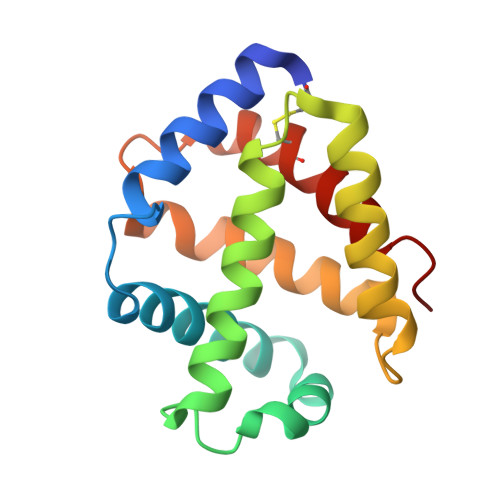
Structures of oxygen dissociation intermediates of 400 kDa V2 hemoglobin provide coarse snapshots of the protein allostery.
Numoto, N., Onoda, S., Kawano, Y., Okumura, H., Baba, S., Fukumori, Y., Miki, K., Ito, N.(2022) Biophys Physicobiol 19: 1-10
- PubMed: 35797404
- DOI: https://doi.org/10.2142/biophysico.bppb-v19.0019
- Primary Citation of Related Structures:
7VLC, 7VLD, 7VLE, 7VLF - PubMed Abstract:
Ever since the historic discovery of the cooperative oxygenation of its multiple subunits, hemoglobin (Hb) has been among the most exhaustively studied allosteric proteins. However, the lack of structural information on the intermediates between oxygenated and deoxygenated forms prevents our detailed understanding of the molecular mechanism of its allostery. It has been difficult to prepare crystals of intact oxy-deoxy intermediates and to individually identify the oxygen saturation for each subunit. However, our recent crystallographic studies have demonstrated that giant Hbs from annelids are suitable for overcoming these problems and can provide abundant information on oxy-deoxy intermediate structures. Here, we report the crystal structures of oxy-deoxy intermediates of a 400 kDa Hb (V2Hb) from the annelid Lamellibrachia satsuma , following up on a series of previous studies of similar giant Hbs. Four intermediate structures had average oxygen saturations of 78%, 69%, 55%, and 26%, as determined by the occupancy refinement of the bound oxygen based on ambient temperature factors. The structures demonstrate that the cooperative oxygen dissociation is weaker, large ternary and quaternary changes are induced at a later stage of the oxygen dissociation process, and the ternary and quaternary changes are smaller with local perturbations. Nonetheless, the overall structural transition seemed to proceed in the manner of the MWC two-state model. Our crystallographic snapshots of the allosteric transition of V2Hb provide important experimental evidence for a more detailed understanding of the allostery of Hbs by extension of the Monod-Wyman-Changeux (MWC) model.
- Medical Research Institute, Tokyo Medical and Dental University (TMDU), Bunkyo-ku, Tokyo 113-8510, Japan.
Organizational Affiliation: