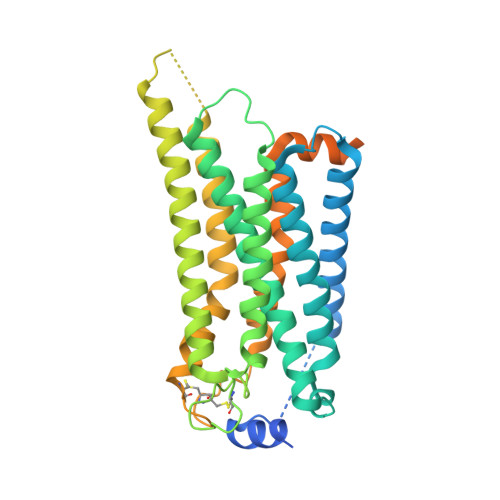
Structural insights into sphingosine-1-phosphate receptor activation.
Yu, L., He, L., Gan, B., Ti, R., Xiao, Q., Hu, H., Zhu, L., Wang, S., Ren, R.(2022) Proc Natl Acad Sci U S A 119: e2117716119-e2117716119
- PubMed: 35412894
- DOI: https://doi.org/10.1073/pnas.2117716119
- Primary Citation of Related Structures:
7VIE, 7VIF, 7VIG, 7VIH - PubMed Abstract:
As a critical sphingolipid metabolite, sphingosine-1-phosphate (S1P) plays an essential role in immune and vascular systems. There are five S1P receptors, designated as S1PR1 to S1PR5, encoded in the human genome, and their activities are governed by endogenous S1P, lipid-like S1P mimics, or nonlipid-like therapeutic molecules. Among S1PRs, S1PR1 stands out due to its nonredundant functions, such as the egress of T and B cells from the thymus and secondary lymphoid tissues, making it a potential therapeutic target. However, the structural basis of S1PR1 activation and regulation by various agonists remains unclear. Here, we report four atomic resolution cryo-electron microscopy (cryo-EM) structures of Gi-coupled human S1PR1 complexes: bound to endogenous agonist d18:1 S1P, benchmark lipid-like S1P mimic phosphorylated Fingolimod [(S)-FTY720-P], or nonlipid-like therapeutic molecule CBP-307 in two binding modes. Our results revealed the similarities and differences of activation of S1PR1 through distinct ligands binding to the amphiphilic orthosteric pocket. We also proposed a two-step “shallow to deep” transition process of CBP-307 for S1PR1 activation. Both binding modes of CBP-307 could activate S1PR1, but from shallow to deep transition may trigger the rotation of the N-terminal helix of Gαi and further stabilize the complex by increasing the Gαi interaction with the cell membrane. We combine with extensive biochemical analysis and molecular dynamic simulations to suggest key steps of S1P binding and receptor activation. The above results decipher the common feature of the S1PR1 agonist recognition and activation mechanism and will firmly promote the development of therapeutics targeting S1PRs.
- Kobilka Institute of Innovative Drug Discovery, School of Life and Health Sciences, The Chinese University of Hong Kong, Shenzhen 518172, China.
Organizational Affiliation: