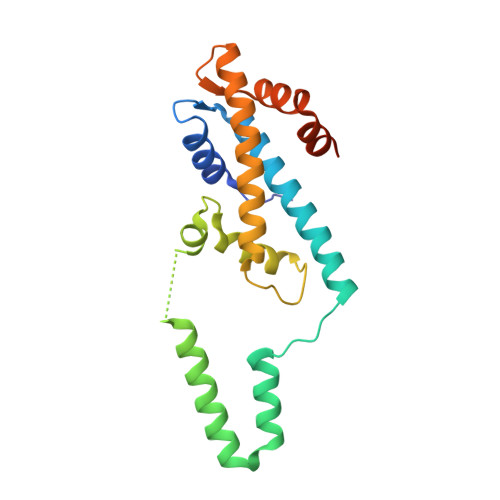
Epstein-Barr Virus Tegument Protein BKRF4 is a Histone Chaperone.
Liu, Y., Li, Y., Bao, H., Liu, Y., Chen, L., Huang, H.(2022) J Mol Biology 434: 167756-167756
- PubMed: 35870648
- DOI: https://doi.org/10.1016/j.jmb.2022.167756
- Primary Citation of Related Structures:
7VCL, 7VCQ - PubMed Abstract:
Histone chaperones, which constitute an interaction and functional network involved in all aspects of histone metabolism, have to date been identified only in eukaryotes. The Epstein-Barr virus tegument protein BKRF4 is a histone-binding protein that engages histones H2A-H2B and H3-H4, and cellular chromatin, inhibiting the host DNA damage response. Here, we identified BKRF4 as a bona fide viral histone chaperone whose histone-binding domain (HBD) forms a co-chaperone complex with the human histone chaperone ASF1 in vitro. We determined the crystal structures of the quaternary complex of the BKRF4 HBD with human H3-H4 dimer and the histone chaperone ASF1b and the ternary complex of the BKRF4 HBD with human H2A-H2B dimer. Through structural and biochemical studies, we elucidated the molecular basis for H3-H4 and H2A-H2B recognition by BKRF4. We also revealed two conserved motifs, D/EL and DEF/Y/W, within the BKRF4 HBD, which may represent common motifs through which histone chaperones target H3-H4 and H2A-H2B, respectively. In conclusion, our results identify BKRF4 as a histone chaperone encoded by the Epstein-Barr virus, representing a typical histone chaperone found in a non-eukaryote. We envision that more histone chaperones await identification and characterization in DNA viruses and even archaea.
- Key Laboratory of Molecular Design for Plant Cell Factory of Guangdong Higher Education Institutes, Department of Biology, School of Life Sciences, Southern University of Science and Technology, Shenzhen 518055, China. Electronic address: yongrui@mail.ustc.edu.cn.
Organizational Affiliation: