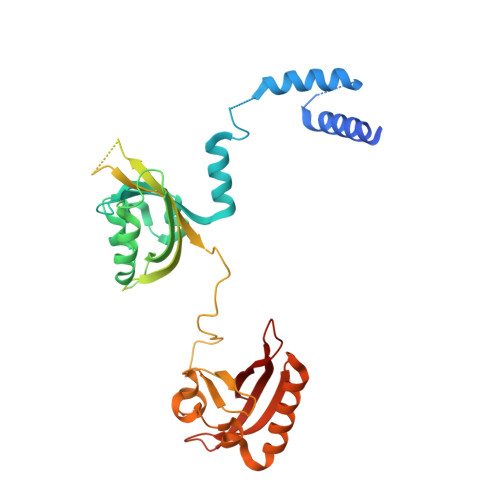
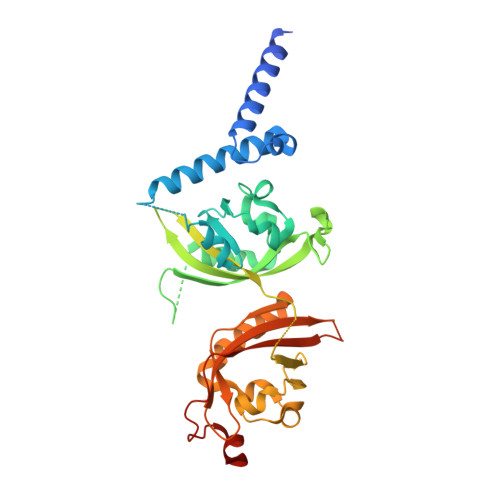
Identification of oleoylethanolamide as an endogenous ligand for HIF-3 alpha.
Diao, X., Ye, F., Zhang, M., Ren, X., Tian, X., Lu, J., Sun, X., Hou, Z., Chen, X., Li, F., Zhuang, J., Ding, H., Peng, C., Rastinejad, F., Luo, C., Wu, D.(2022) Nat Commun 13: 2529-2529
- PubMed: 35534502
- DOI: https://doi.org/10.1038/s41467-022-30338-z
- Primary Citation of Related Structures:
7V7L, 7V7W - PubMed Abstract:
Hypoxia-inducible factors (HIFs) are α/β heterodimeric transcription factors modulating cellular responses to the low oxygen condition. Among three HIF-α isoforms, HIF-3α is the least studied to date. Here we show that oleoylethanolamide (OEA), a physiological lipid known to regulate food intake and metabolism, binds selectively to HIF-3α. Through crystallographic analysis of HIF-3 α/β heterodimer in both apo and OEA-bound forms, hydrogen-deuterium exchange mass spectrometry (HDX-MS), molecular dynamics (MD) simulations, and biochemical and cell-based assays, we unveil the molecular mechanism of OEA entry and binding to the PAS-B pocket of HIF-3α, and show that it leads to enhanced heterodimer stability and functional modulation of HIF-3. The identification of HIF-3α as a selective lipid sensor is consistent with recent human genetic findings linking HIF-3α with obesity, and demonstrates that endogenous metabolites can directly interact with HIF-α proteins to modulate their activities, potentially as a regulatory mechanism supplementary to the well-known oxygen-dependent HIF-α hydroxylation.
- Helmholtz International Lab, State Key Laboratory of Microbial Technology, Shandong University, 266237, Qingdao, China.
Organizational Affiliation: