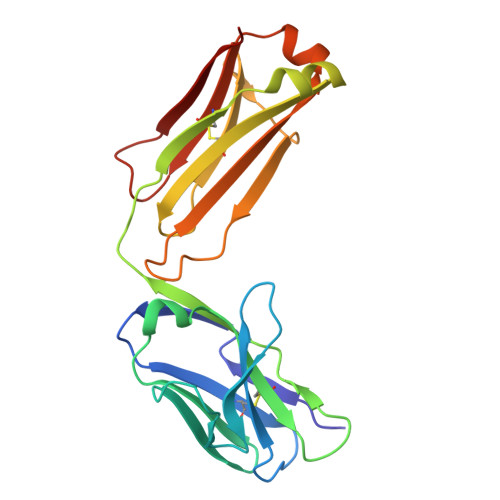
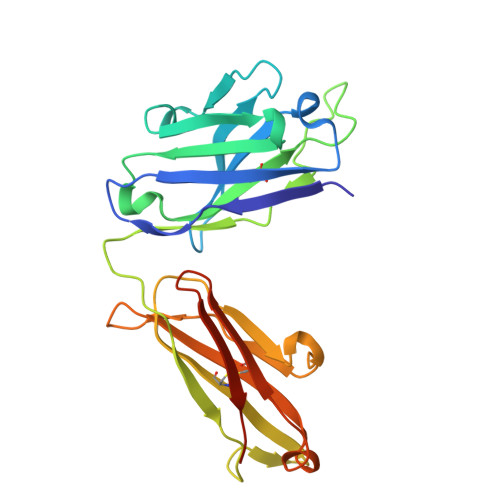
Development of a DNA aptamer that binds to the complementarity-determining region of therapeutic monoclonal antibody and affinity improvement induced by pH-change for sensitive detection.
Saito, T., Shimizu, Y., Tsukakoshi, K., Abe, K., Lee, J., Ueno, K., Asano, R., Jones, B.V., Yamada, T., Nakano, T., Tong, J., Hishiki, A., Hara, K., Hashimoto, H., Sode, K., Toyo'oka, T., Todoroki, K., Ikebukuro, K.(2022) Biosens Bioelectron 203: 114027-114027
- PubMed: 35114463
- DOI: https://doi.org/10.1016/j.bios.2022.114027
- Primary Citation of Related Structures:
7V5N - PubMed Abstract:
Therapeutic monoclonal antibodies (mAbs) are successful biomedicines; however, evaluation of their pharmacokinetics and pharmacodynamics demands highly specific discrimination from human immunoglobulin G naturally present in the blood. Here, we developed a novel anti-idiotype aptamer (termed A14#1) with extraordinary specificity against the anti-vascular endothelial growth factor therapeutic mAb, bevacizumab. Structural analysis of the antibody-aptamer complex showed that several bases of A14#1 recognized only the complementarity determining region (CDR) of bevacizumab, thereby contributing to its extraordinary specificity. As the CDR of bevacizumab is predicted to be highly positively charged under mildly acidic conditions and that DNA is negatively charged, the affinity of A14#1 to bevacizumab markedly increased at pH 4.7 (K D = 44 pM) than at pH 7.4 (K D = 12 nM). A14#1-based electrochemical detection method capable of detecting 31 pM of bevacizumab at pH 4.7 was thus developed. A14#1 could be potentially useful for therapeutic drug measurement as a novel ligand of bevacizumab.
- Department of Biotechnology and Life Science, Tokyo University of Agriculture and Technology, 2-24-16 Naka-cho, Koganei, Tokyo, 184-8588, Japan.
Organizational Affiliation: