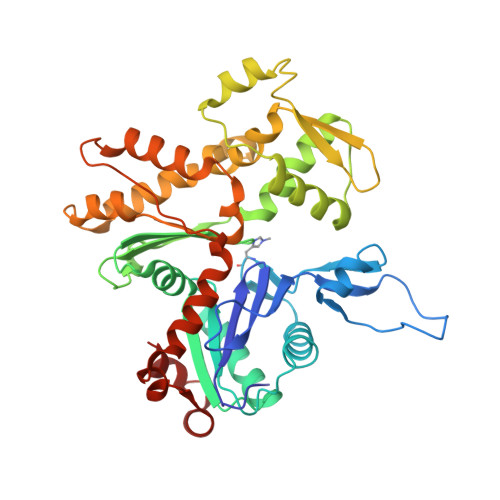
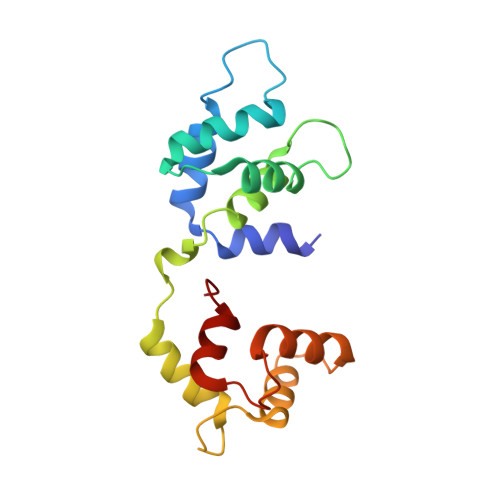
Protein-Protein Docking Reveals Dynamic Interactions of Tropomyosin on Actin Filaments.
Pavadai, E., Lehman, W., Rynkiewicz, M.J.(2020) Biophys J 119: 75-86
- PubMed: 32521240
- DOI: https://doi.org/10.1016/j.bpj.2020.05.017
- Primary Citation of Related Structures:
7UTI, 7UTL - PubMed Abstract:
Experimental approaches such as fiber diffraction and cryo-electron microscopy reconstruction have defined regulatory positions of tropomyosin on actin but have not, as yet, succeeded at determining key atomic-level contacts between these proteins or fully substantiated the dynamics of their interactions at a structural level. To overcome this deficiency, we have previously employed computational approaches to deduce global dynamics of thin filament components by energy landscape determination and molecular dynamics simulations. Still, these approaches remain computationally challenging for any complex and large macromolecular assembly like the thin filament. For example, tropomyosin cable wrapping around actin of thin filaments features both head-to-tail polymeric interactions and local twisting, both of which depart from strict superhelical symmetry. This produces a complex energy surface that is difficult to model and thus to evaluate globally. Therefore, at this stage of our understanding, assessing global molecular dynamics can prove to be inherently impractical. As an alternative, we adopted a "divide and conquer" protocol to investigate actin-tropomyosin interactions at an atomistic level. Here, we first employed unbiased protein-protein docking tools to identify binding specificity of individual tropomyosin pseudorepeat segments over the actin surface. Accordingly, tropomyosin "ligand" segments were rotated and translated over potential "target" binding sites on F-actin where the corresponding interaction energetics of billions of conformational poses were ranked by the programs PIPER and ClusPro. These data were used to assess favorable interactions and then to rebuild models of seamless and continuous tropomyosin cables over the F-actin substrate, which were optimized further by flexible fitting routines and molecular dynamics. The models generated azimuthally distinct regulatory positions for tropomyosin cables along thin filaments on actin dominated by stereo-specific head-to-tail overlap linkage. The outcomes are in good agreement with current cryo-electron microscopy topology and consistent with long-thought residue-to-residue interactions between actin and tropomyosin.
- Department of Physiology & Biophysics, Boston University School of Medicine, Boston, Massachusetts.
Organizational Affiliation: