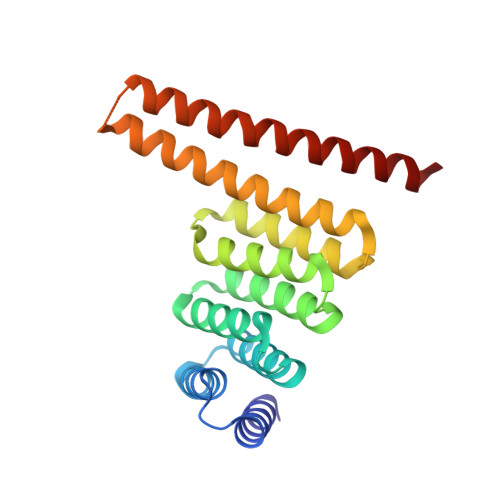
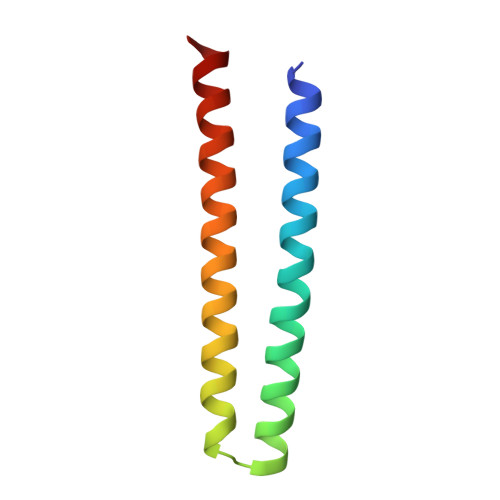
De novo design of obligate ABC-type heterotrimeric proteins.
Bermeo, S., Favor, A., Chang, Y.T., Norris, A., Boyken, S.E., Hsia, Y., Haddox, H.K., Xu, C., Brunette, T.J., Wysocki, V.H., Bhabha, G., Ekiert, D.C., Baker, D.(2022) Nat Struct Mol Biol 29: 1266-1276
- PubMed: 36522429
- DOI: https://doi.org/10.1038/s41594-022-00879-4
- Primary Citation of Related Structures:
7UPO, 7UPP, 7UPQ - PubMed Abstract:
The de novo design of three protein chains that associate to form a heterotrimer (but not any of the possible two-chain heterodimers) and that can drive the assembly of higher-order branching structures is an important challenge for protein design. We designed helical heterotrimers with specificity conferred by buried hydrogen bond networks and large aromatic residues to enhance shape complementary packing. We obtained ten designs for which all three chains cooperatively assembled into heterotrimers with few or no other species present. Crystal structures of a helical bundle heterotrimer and extended versions, with helical repeat proteins fused to individual subunits, showed all three chains assembling in the designed orientation. We used these heterotrimers as building blocks to construct larger cyclic oligomers, which were structurally validated by electron microscopy. Our three-way junction designs provide new routes to complex protein nanostructures and enable the scaffolding of three distinct ligands for modulation of cell signaling.
- Department of Biochemistry, University of Washington, Seattle, WA, USA.
Organizational Affiliation: