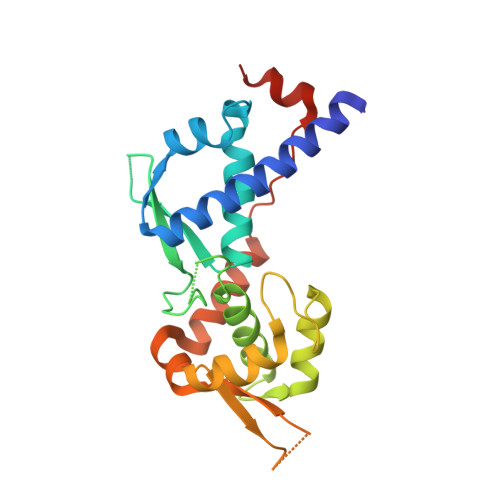
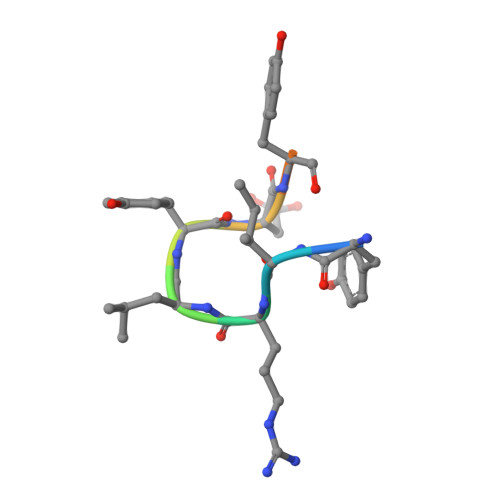
Discovery and Structural Basis of the Selectivity of Potent Cyclic Peptide Inhibitors of MAGE-A4.
Fleming, M.C., Chiou, L.F., Tumbale, P.P., Droby, G.N., Lim, J., Norris-Drouin, J.L., Williams, J.G., Pearce, K.H., Williams, R.S., Vaziri, C., Bowers, A.A.(2022) J Med Chem 65: 7231-7245
- PubMed: 35522528
- DOI: https://doi.org/10.1021/acs.jmedchem.2c00185
- Primary Citation of Related Structures:
7UOA - PubMed Abstract:
MAGE proteins are cancer testis antigens (CTAs) that are characterized by highly conserved MAGE homology domains (MHDs) and are increasingly being found to play pivotal roles in promoting aggressive cancer types. MAGE-A4, in particular, increases DNA damage tolerance and chemoresistance in a variety of cancers by stabilizing the E3-ligase RAD18 and promoting trans-lesion synthesis (TLS). Inhibition of the MAGE-A4:RAD18 axis could sensitize cancer cells to chemotherapeutics like platinating agents. We use an mRNA display of thioether cyclized peptides to identify a series of potent and highly selective macrocyclic inhibitors of the MAGE-A4:RAD18 interaction. Co-crystal structure indicates that these inhibitors bind in a pocket that is conserved across MHDs but take advantage of A4-specific residues to achieve high isoform selectivity. Cumulatively, our data represent the first reported inhibitor of the MAGE-A4:RAD18 interaction and establish biochemical tools and structural insights for the future development of MAGE-A4-targeted cellular probes.
- Division of Chemical Biology and Medicinal Chemistry, Eshelman School of Pharmacy, University of North Carolina, Chapel Hill, North Carolina 27599, United States.
Organizational Affiliation: