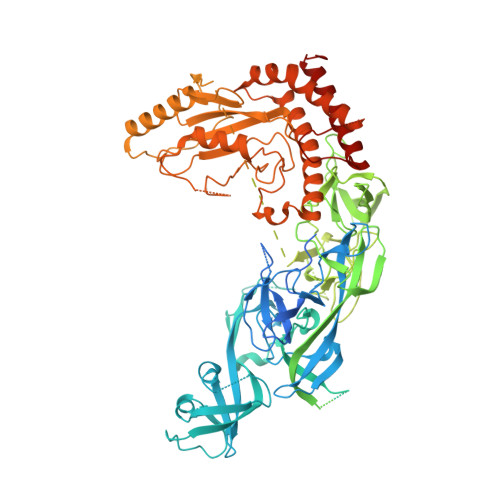
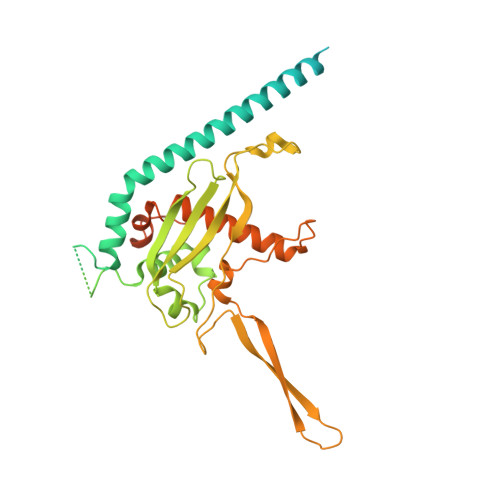
Mechanism of client selection by the protein quality-control factor UBE2O.
Yip, M.C.J., Sedor, S.F., Shao, S.(2022) Nat Struct Mol Biol 29: 774-780
- PubMed: 35915257
- DOI: https://doi.org/10.1038/s41594-022-00807-6
- Primary Citation of Related Structures:
7UN3, 7UN6 - PubMed Abstract:
The E2/E3 enzyme UBE2O ubiquitylates diverse clients to mediate important processes, including targeting unassembled 'orphan' proteins for quality control and clearing ribosomes during erythropoiesis. How quality-control factors, such as UBE2O, select clients on the basis of heterogeneous features is largely unknown. Here, we show that UBE2O client selection is regulated by ubiquitin binding and a cofactor, NAP1L1. Attaching a single ubiquitin onto a client enhances UBE2O binding and multi-mono-ubiquitylation. UBE2O also repurposes the histone chaperone NAP1L1 as an adapter to recruit a subset of clients. Cryo-EM structures of human UBE2O in complex with NAP1L1 reveal a malleable client recruitment interface that is autoinhibited by the intrinsically reactive UBC domain. Adding a ubiquitylated client identifies a distinct ubiquitin-binding SH3-like domain required for client selection. Our findings reveal how multivalency and a feed-forward mechanism drive the selection of protein quality-control clients.
- Department of Cell Biology, Harvard Medical School, Boston, MA, USA.
Organizational Affiliation: