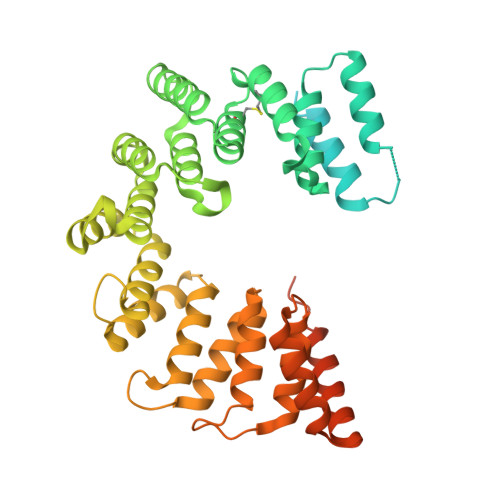
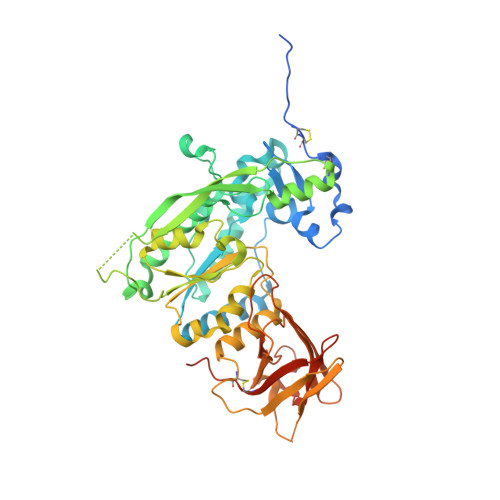
Structure of the AlgKX modification and secretion complex required for alginate production and biofilm attachment in Pseudomonas aeruginosa.
Gheorghita, A.A., Li, Y.E., Kitova, E.N., Bui, D.T., Pfoh, R., Low, K.E., Whitfield, G.B., Walvoort, M.T.C., Zhang, Q., Codee, J.D.C., Klassen, J.S., Howell, P.L.(2022) Nat Commun 13: 7631-7631
- PubMed: 36494359
- DOI: https://doi.org/10.1038/s41467-022-35131-6
- Primary Citation of Related Structures:
7ULA - PubMed Abstract:
Synthase-dependent secretion systems are a conserved mechanism for producing exopolysaccharides in Gram-negative bacteria. Although widely studied, it is not well understood how these systems are organized to coordinate polymer biosynthesis, modification, and export across both membranes and the peptidoglycan. To investigate how synthase-dependent secretion systems produce polymer at a molecular level, we determined the crystal structure of the AlgK-AlgX (AlgKX) complex involved in Pseudomonas aeruginosa alginate exopolysaccharide acetylation and export. We demonstrate that AlgKX directly binds alginate oligosaccharides and that formation of the complex is vital for polymer production and biofilm attachment. Finally, we propose a structural model for the AlgEKX outer membrane modification and secretion complex. Together, our study provides insight into how alginate biosynthesis proteins coordinate production of a key exopolysaccharide involved in establishing persistent Pseudomonas lung infections.
- Program in Molecular Medicine, The Hospital for Sick Children, Toronto, ON, Canada.
Organizational Affiliation: